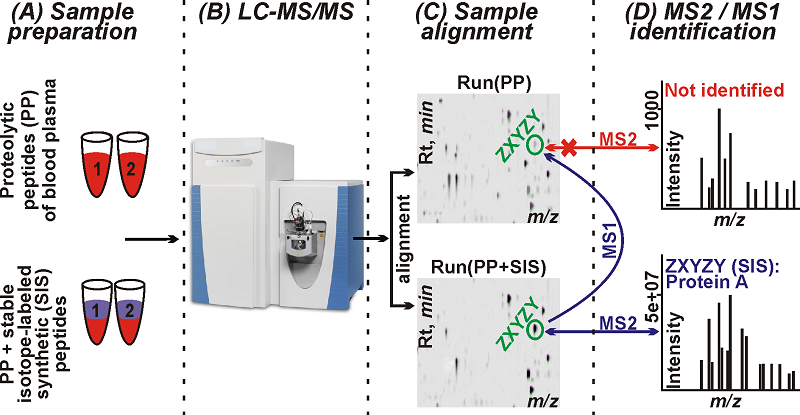

Figure 1. Diagram of an experiment allowing workflow co-alignment of samples of proteolytic peptides of human blood plasma with and without spike-in synthetic peptides. The spike-in peptides were aligned with the natural peptides by the RTs and m/z of their MS1 features in order to identify proteins by high quality MS2 spectra of spike-in synthetic peptides. Key stages: (A) sample preparation; (B) LC-MS/MS analysis by high resolution mass spectrometer; (C) the samples alignment using Progenesis software; (D) search for protein identifications in Mascot software; mapping of MS2 synthetic peptide identifications to MS1 features of aligned samples and the identification of human blood plasma proteins corresponding to spike-in peptides in a sample free of these peptides.