Biomedical Chemistry: Research and Methods
2022, 5(1), e00164
The Study of Sensitivity to Proteolysis of Full-Length and Truncated Forms of Recombinant Human Renalase Expressed in the prokaryotic System
Institute of Biomedical Chemistry, 10 Pogodinskaya str., Moscow, 119121 Russia; *e-mail: valfed38@yandex.ru
Keywords: renalase, trypsin, proteolysis
DOI:10.18097/BMCRM00164
The whole version of this paper is available in Russian.
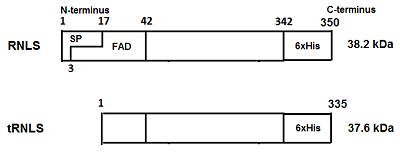
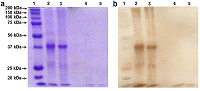
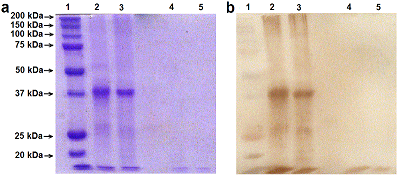
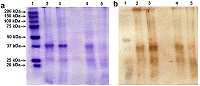
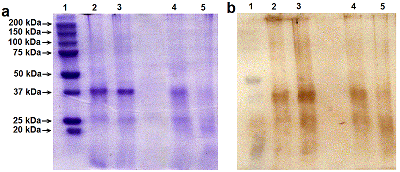
Renalase (RNLS) is a flavoprotein; its N-terminal peptide (amino acid residues 1-17) performs various important functions. Inside cells, it is involved in the Rossmann fold formation (residues 2-35), which is necessary for the binding of the FAD cofactor and the manifestation of the enzymatic activity of RNLS as a FAD-dependent oxidoreductase (EC 1.6.3.5). When RNLS is secreted into the extracellular space, this peptide is cleaved off, and the resulting truncated extracellular RNLS can no longer bind FAD and, therefore, numerous effects described in the literature are carried out by non-catalytic mechanisms. In this work, we have investigated the sensitivity to trypsinolysis of two recombinant forms of human RNLS expressed in prokaryotic cells: (a) full-length RNLS containing the FAD cofactor; (b) a truncated RNLS lacking the 1-17 N-terminal peptide (truncatedRNLS, tRNLS) unable to bind the FAD cofactor. Trypsin (1 unit/20 μL of medium) effectively cleaved both forms of renalase (RNLS and tRNLS). When exposed to a lower concentration of trypsin (0.1 U/20 µL of medium), full length RNLS was more trypsin resistant than tRNLS. We suggest that the different sensitivity of RNLS and tRNLS is apparently determined by the presence of the FAD cofactor in the full-length recombinant protein, which contributes to the formation of a spatial structure that is more resistant to the action of certain proteases.
FUNDING
This work was supported by the Russian Foundation for Basic Research (project no. 20–015–00104).
REFERENCES
- Xu, J., Li G., Wang, P., Velazquez, H., Yao, X., Li, Y., Wu, Y., Peixoto, A., Crowley, S., Desir, G.V. (2005) Renalase is a novel, soluble monoamine oxidase that regulates cardiac function and blood pressure, J. Clin. Invest., 115, 1275-1280. DOI
- Moran, G.R., Hoag, M.R. (2017) The enzyme: Renalase, Arch Biochem Biophys., 632, 66-76. DOI
- Fedchenko, V., Kopylov, A., Kozlova, N., Buneeva, O., Kaloshin, A., Zgoda, V., Medvedev, A. (2016) Renalase secreted by human kidney НЕК293Т cells lacks its N-terminal peptide: implications for putative mechanisms of renalase action. Kidney Blood Pressure Research, 41, 593-603
- Pointer, T.C., Gorelick, F.S., Desir, G.V. (2021) Renalase: A Multi-Functional Signaling Molecule with Roles in Gastrointestinal Disease, Cells, 10, 2006. DOI
- Kolodecik, T.R., Reed, A.M., Date, K., Shugrue, C.A., Patel, V., Chung, S.L., Desir, G.V., Gorelick, F.S. (2017) The serum protein renalase reduces injury in experimental pancreatitis. J. Biol. Chem., 292, 21047-21059. DOI
- Wang, L., Velazquez, H., Chang, J., Safirstein, R., Desir, G.V. (2015) Identification of a receptor for extracellular renalase.PLoS One. 10, e0122932. DOI
- Wang, Y., Safirstein, R., Velazquez, H., Guo, X.J., Hollander, L., Chang, J., Chen, T.M., Mu, J.J., Desir, G.V.(2017) Extracellular renalase protects cells and organs by outside-in signalling. J. Cell. Mol. Med., 21, 1260-1265. DOI
- Medvedev, A., Kopylov, A., Fedchenko, V., Buneeva, O. (2020) Is renalase ready to become a biomarker of ischemia? Int. J. Cardiol. 307, 179. DOI
- Fedchenko, V. I., Kaloshin, A.A., Mezhevikina, L.M., Buneeva, O.A., Medvedev, A.E. (2013) Construction of the coding sequence of the transcription variant 2 of the human renalase gene and its expression in the prokaryotic system. Int. J. Mol. Sci. 14, 12764-12779. DOI
- Fedchenko, V.I., Kaloshin, A.A. (2019) A simplified method for obtaining cDNA of low-copy and silent eukaryotic genes using human renalase as anexample. Biomedical Chemistry: Research and Methods. 2(2), e00101. DOI
- Fedchenko, V.I., Kaloshin, A.A., Kozlova, N.I., Kopylov, A.T., Medvedev, A.E.(2020) Construction of a chimeric human gene encoding renalase with a modified N-terminus. Biomedical Chemistry: Research and Methods, 3(3), e00137. DOI
- Fedchenko, V.I., Kaloshin, A.A., Kaloshina,, S.A, Medvedev, A.E.(2021) Expression and isolation of N-terminal truncated human recombinant renalasein prokaryotic cells. Biomedical Chemistry: Research and Methods , 4, e00158 DOI
- Laemmli, U.K. (1970) Cleavage of structural proteins during the assemblyof the head of bacteriophage T4. Nature; 227, 680-685. DOI
- Gallagher, S., Winston, ,S.E., Fuller, S.A., Hurrell, J.G.R. (2011) Immunoblotting and immunodetection. Curr.Protoc. Cell Biol. 52, 6.2.1-6.2.28. DOI