Bigdata of National Medicine Registers
Institute of Biomedical Chemistry, 10 Pogodinskaya str., Moscow, 119121 Russia; *e-mail: polina.savosina@ibmc.msk.ru
Keywords: BigData; approved drugs; national medicine registers; medicine agencies; web-resources; World Wide Approved Drugs
DOI:10.18097/BMCRM00230
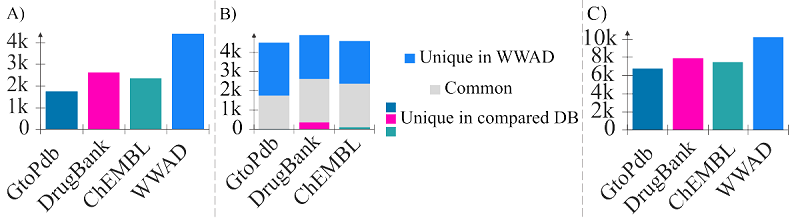
Currently, various databases of approved drugs are widely used in the development and application of computer-aided drug design for training sets and validating predicted models. Most of the freely available databases contain information on a limited number of drugs. This leads to reduction in the space of pharmacotherapeutic chemistry studied and used by researchers. Information on drugs developed and used locally, in one or two countries, can be obtained from relevant national medicine registries. We have identified and analyzed registries from more than 70 countries around the world that are accessible through open web resources on the Internet. In addition to lists of approved therapeutics, many web resources offer the opportunity to review official documents published by medical authorities after the approval process. These documents contain a wide range of information about drugs, including data on structural formulations, pharmacodynamics and pharmacokinetics, toxicity, etc. The compounds represented in the registries, both in terms of quantity and structural diversity, exceed the known widely used databases of approved drugs. Combined data from national drug registries represent an example of "Big Data" in the biomedical field, taking into account all the difficulties involved in their processing. Its use in computer-aided drug design will not only expand the pharmacotherapeutic chemical space studied, but also improve the quality of the original data.

|
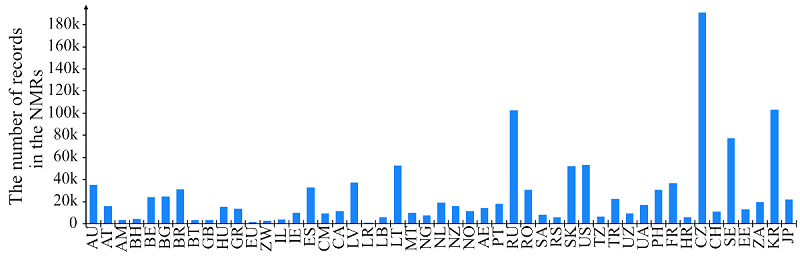
Figure 1.
The number of records in the found 49 national medicine registers. Here and further the country names are given in the ISO 3166-1 system codes.
|

|
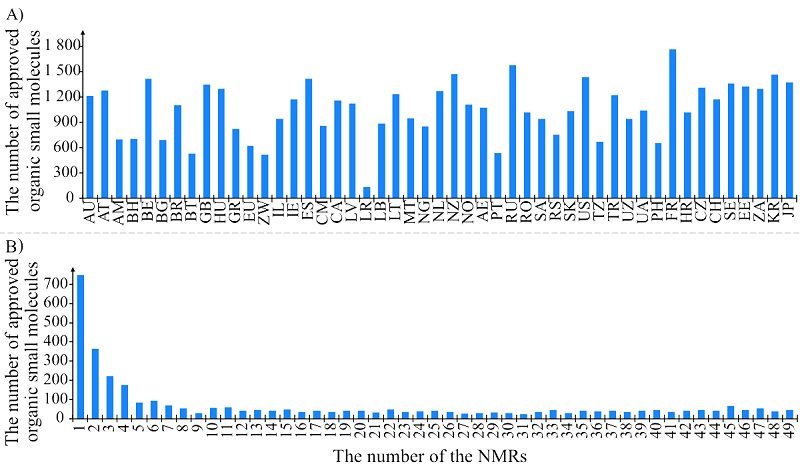
Figure 3.
The number of unique organic low molecular weight drug compounds: A) in the 49 analyzed NMRs; B) depending on the number of the NMRs in which these substances are presented.
|
FUNDING
The work was performed within the framework of the Program for Basic Research in the Russian Federation for a long-term period (2021–2030) (No. 122030100170-5).
REFERENCES
- Tanoli, Z., Seemab, U., Scherer, A., Wennerberg, K., Tang, J., Vähä-Koskela, M. (2021) Exploration of databases and methods supporting drug repurposing: A comprehensive survey. Briefings Bioinformatics, 22(2), 1656–1678. DOI
- Siramshetty, V.B., Grishagin, I., Nguyễn, Ð.T., Peryea, T., Skovpen, Y., Stroganov, O., Katzel, D., Sheils, T., Jadhav, A., Mathé, E.A., Southall, N.T. (2022) NCATS Inxight Drugs: A comprehensive and curated portal for translational research. Nucleic Acids Res., 50(D1), D1307–D1316. DOI
- Maglo, K.N., Mersha, T.B., Martin, L.J. (2016) Population genomics and the statistical values of race: An interdisciplinary perspective on the biological classification of human populations and implications for clinical genetic epidemiological research. Front. Genet., 7, 22. DOI
- World Health Organization. Competent authorities of countries participating in the WHO. Retrieved June 11, 2024, from: https://www.who.int/teams/regulation-prequalification/regulation-and-safety/regulatory-convergence-networks/certification-scheme/contacts.
- Leaman, R., Islamaj, R., Adams, V., Alliheedi, M.A.,Almeida, J.R., Antunes, R., Bevan, R., Chang, Y.C., Erdengasileng, A., Hodgskiss, M., Ida, R., Kim, H., Li, K., Mercer, R.E., Mertová, L., Mobasher, G., Shin, H.C., Sung, M., Tsujimura, T., Yeh, W.C., Lu, Z. (2023) Chemical identification and indexing in full-text articles: An overview of the NLM-Chem track at BioCreative VII. Database, 2023, baad005. DOI
- Chaudhuri, A., Mandaviya, K., Badelia, P., Ghosh, S.K. (2017) Summary and Future Research. In: Optical Character Recognition Systems for Different Languages with Soft Computing. Studies in Fuzziness and Soft Computing (Chaudhuri A., Mandaviya K., Badelia P., Ghosh S.K., eds.) Springer: Cham, Switzerland, pp. 241–245.
- World Wide Approved Drugs database. Retrieved June 11, 2024, from: https://www.way2drug.com/wwad.
- DrugBank database. Approved drugs. Retrieved June 11, 2024, from: https://go.drugbank.com/drugs.
- ChEMBL database. Approved drugs. Retrieved June 11, 2024, from: https://www.ebi.ac.uk/chembl.
- IUPHAR/BPS Guide to PHARMACOLOGY database. Approved drugs. Retrieved June 11, 2024, from: https://www.guidetopharmacology.org/GRAC/LigandListForward ?type=Approved.
- Filimonov, D., Druzhilovskiy, D., Lagunin, A., Gloriozova, T., Rudik, A., Dmitriev, A., Pogodin, P., Poroikov, V. (2018) Computer-aided prediction of biological activity spectra for chemical compounds: Opportunities and limitations. Biomedical Chemistry: Research and Methods, 1(1), e00004. DOI