Modeling the Blood-Brain Barrier Using Rat Brain Cell Cultures
Institute of Biomedical Chemistry, 10 Pogodinskaya str., Moscow, 119121 Russia; *e-mail: roza_saryglar@mail.ru
Keywords: blood-brain barrier, endothelial cells, astrocytes, pericytes, cell co-cultivation
DOI:10.18097/BMCRM00238
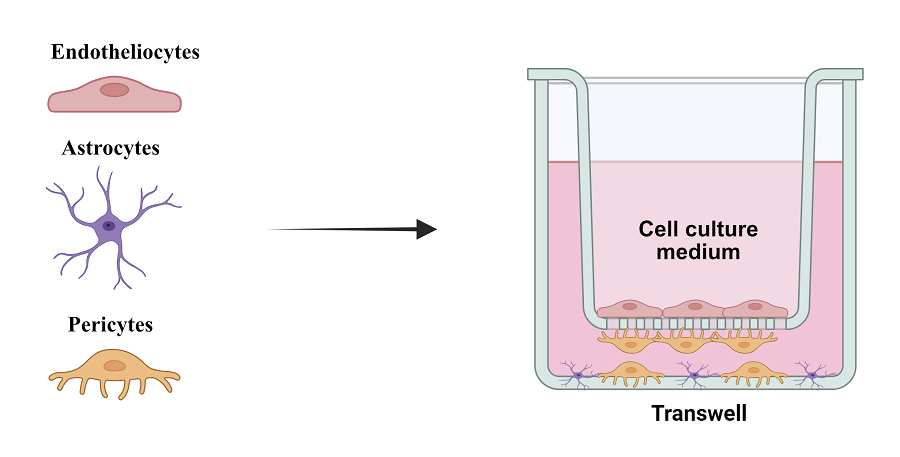
Permeability of the blood-brain barrier (BBB) represents a significant problem for most promising drugs used for treating brain diseases due to its high selectivity. The neurovascular unit, which includes neurons, interneurons, astrocytes, the basal membrane, smooth muscle cells, pericytes, endothelial cells, and the extracellular matrix, forms an anatomically and functionally cohesive structure that ensures effective regulation of cerebral blood flow. In vitro modeling of the BBB is a relevant and practically significant task for studying the penetration of therapeutic agents into the brain. This study presents a BBB model consisting of endothelial cells, pericytes, and astrocytes, which partially mimics the in vivo layers of the BBB. Despite some limitations, such as incomplete matching of astrocyte location, the model demonstrates high expression of tight junction proteins and optimal TEER values of the endothelial cell monolayer, making it suitable for studying the permeability of the BBB to various substances, including drugs and nanoparticles.

|
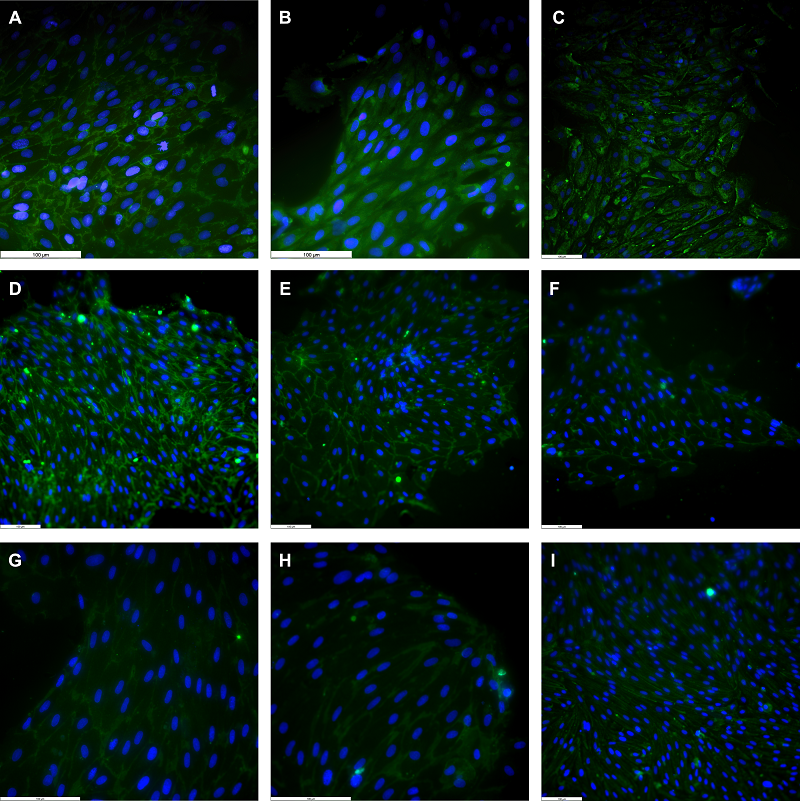
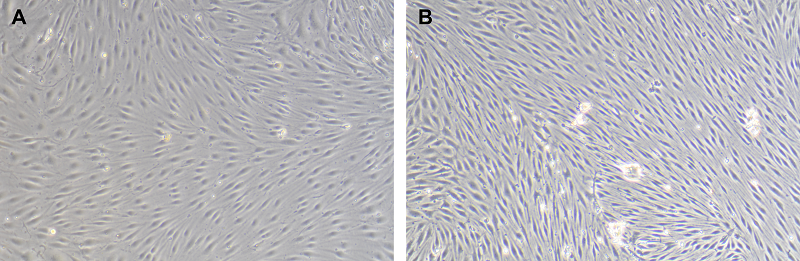
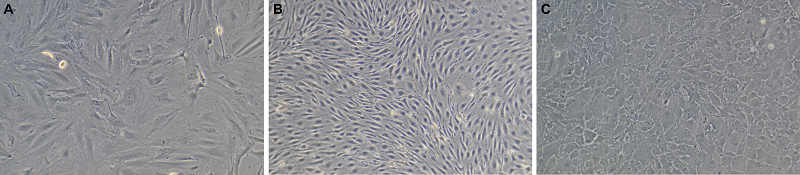
Figure 1.
Phase-contrast images of primary cell cultures isolated from rat brain. A – pericytes, B – endotheliocytes, C – astrocytes. Magnification X100.
|


|
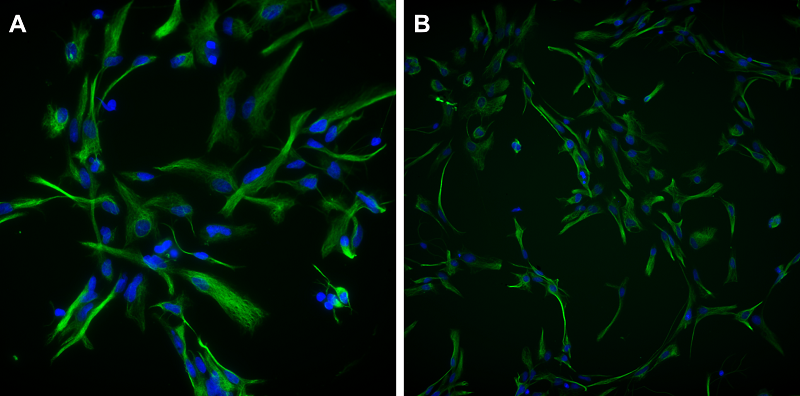
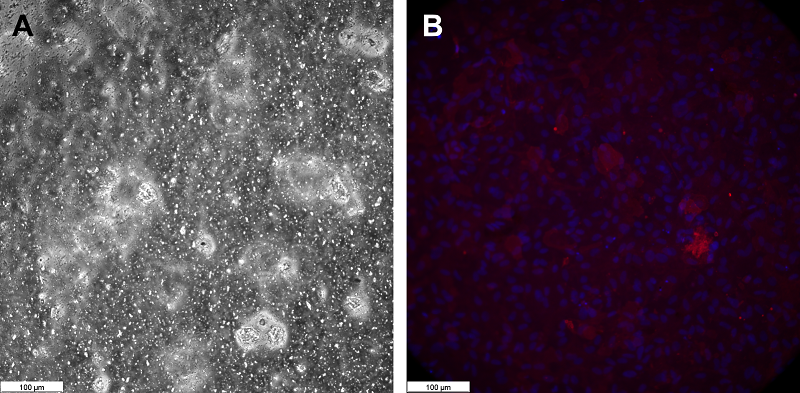
Figure 2.
αSMA expression in pericytes isolated from rat brain microvessels. Fluorescence microscopy. DAPI is used to stain cell nuclei. Magnification X100.
|
FUNDING
The study was performed in the framework of the Program for Basic Research in the Russian Federation for long-term period (2021-2030) No. 122022800499-5).
REFERENCES
- Xu, J., Li, G., Wang, P., Velazquez, H., Yao, X., Li, Y., Wu, Y., Peixoto, A., Crowley, S., Desir, G.V. (2005) Renalase is a novel, soluble monoamine oxidase that regulates cardiac function and blood pressure. J. Clin. Invest., 115(5), 1275–1280. DOI
- Severina, I.S., Fedchenko, V.I., Veselovsky, A.V., Medvedev, A.E. (2015)The history of renalase from amine oxidase to a α-NAD(P)H-oxidase/anomerase. Biomeditsinskaya Khimiya. 61(6), 667-679. DOI
- Pointer, T.C., Gorelick, F.S., Desir, G.V. (2021) Renalase: a multi-functional signaling molecule with roles in gastrointestinal disease, Cells, 10, 2006. DOI
- Moran, G.R., Hoag, M.R. (2017) The enzyme: Renalase. Arch. Biochem. Biophys, 632, 66-76. DOI
- Fedchenko, V., Kopylov, A., Kozlova, N., Buneeva, O., Kaloshin, A., Zgoda, V., Medvedev, A. (2016) Renalase secreted by human kidney HEK293T cells lacks its n-terminal peptide: implications for putative mechanisms of renalase action, Kidney Blood Press Res. 41, 593-603. DOI
- Wang, Y., Safirstein, R., Velazquez, H., Guo, X.J., Hollander, L., Chang, J., Chen, T.M., Mu, J.J., Desir, G.V. (2017) Extracellular renalase protects cells and organs by outside-in signalling. J. Cell Mol. Med., 21(7), 1260-1265. DOI
- Kolodecik, T.R., Reed, A.M., Date, K., Shugrue, C.A., Patel, V., Chung, S.L., Desir, G.V., Gorelick, F.S. (2017) The serum protein renalase reduces injury in experimental pancreatitis. J. Biol. Chem., 292(51), 21047–21059. DOI
- Zhan,g L., Zang, C.S., Chen, B., Wang, Y., Xue, S., Wu, M.Y. (2023) Renalase regulates renal tubular injury in diabetic nephropathy via the p38MAPK signaling pathway. FASEB J. 37(10), e23188. DOI
- Retrieved 01.10.2024 from https://www.abcam.com/en-us/products/primary-antibodies/renalase-antibody-ab31291
- Fedchenko, V.I., Kaloshin, A.A., Mezhevikina, L.M., Buneeva, O.A., Medvedev, A.E. (2013) Construction of the coding sequence of the transcription variant 2 of the human Renalase gene and its expression in the prokaryotic system. Int. J. Mol. Sci.,14(6),12764-1279. DOI
- Fedchenko, V.I., Kaloshin, A.A. (2019) A simplified method for obtaining cDNA of low-copy and silent eukaryotic genes using human renalase as an example. Biomedical Chemistry: Research and Methods. 2(2), e00101. DOI
- Fedchenko, V., Kaloshin, A., Medvedev, A. (2023). Improvement of the exon method for rapid synthesis of cDNA of the rat renalase gene. Biomedical Chemistry: Research and Methods, 6(3), e00201. DOI
- Fedchenko, V., Kaloshin, A., Medvedev, A. (2024). Generation of C-terminal sequences of human renalase-1 and renalase-2 encoded by alternative exons. Biomedical Chemistry: Research and Methods, 7(2), e00228. DOI
- Laemmli, U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227(5259), 680-685. DOI
- Gallagher, S., Winston, S.E., Fuller, S.A., Hurrell, J.G. (2008) Immunoblotting and immunodetection. Curr. Protoc. Mol. Biol., Chapter 10:Unit 10.8. DOI
- Fedchenko, V.I., Veselovsky, A.V., Kopylov, A.T., Kaloshina, S.A., Medvedev, A.E. (2022) Renalase may be cleaved in blood. Are blood chymotrypsin-like enzymes involved? Medical Hypotheses, 165, 110895. DOI
- Fedchenko, V., Globa, A., Buneeva, O., Medvedev, A. (2013) Renalase mRNA levels in the brain, heart, and kidneys of spontaneously hypertensive rats with moderate and high hypertension. Med. Sci. Monit. Basic Res. 19, 267-270. DOI
- Retrieved 01.10.2024 from https://blast.ncbi.nlm.nih.gov/Blast.cgi.