The 40th Anniversary of the Institute of Physiologically Active Compounds of the Russian Academy of Sciences
Computational Evaluation of Selectivity of Inhibition of Muscarinic Receptors M1-M4
1Institute of Physiologically Active Compounds of the Russian Academy of Sciences, 1 Severny proezd, Moscow region, Chernogolovka, 142432 Russia,*e-mail: a.v.mikurova@ibmc.msk.ru 2Institute of Biomedical Chemistry, 10 Pogodinskaya Street, Moscow, 119121, Russia
Key words: acetylcholine muscarinic receptors; inhibitors; comparative inhibition; docking; computational methods; molecular
dynamics; QSAR
DOI: 10.18097/BMCRM00072
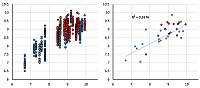
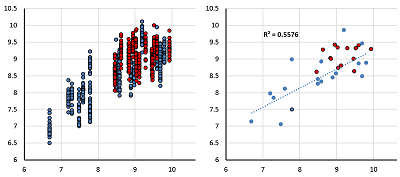
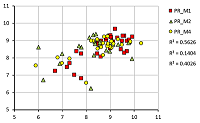
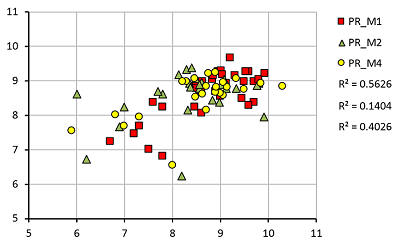
A set of models for preliminary estimation of the inhibition constant values of potential ligands for the 4 acetylcholine muscarinic receptors M1-M4 was developed. The study uses an information about three-dimensional structure of human M1, M2 and M4 receptors, as well as the M3 receptor model, constructed by homology based on the structure of the rat M3 receptor. The Ki values for 42 compounds were obtained from the sources. Modeling of "protein-ligand" complexes was performed using molecular docking and molecular dynamics procedures. The component energy characteristics of the complexes were calculated from data obtained from simulation of molecular dynamics by the MM-PBSA/MM-GBSA methods. These characteristics were used as independent variables to construct the linear regression equations for pKi value predicting. The equations obtained for each receptors allow us to predict pKi with an average accuracy of 0.65 logarithmic units

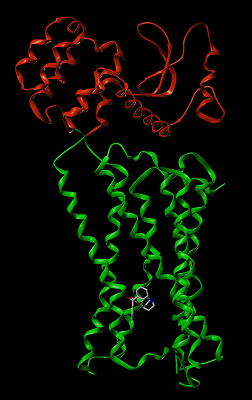
|
Figure 1.
The structure of human M2 acetylcholine muscarinic receptor. Chimeric part is marked by red. The binding site located at a considerable distance from the chimeric part.
|


|
Figure 2.
Comparison of sequences of human and rat acetylcholine muscarinic receptors. Differences are marked by red. The chimeric part is underlined.
|
|
CLOSE

|
Table 1.
Strucrures of acetylcholine muscarinic receptors available in PDB.
|
|
CLOSE

|
Table 2.
Antagonists of acetylcholine muscarinic receptors.
|
|
CLOSE

|
Table 3.
Learning parameters for linear regression models for particular receptors.
|
|
CLOSE

|
Table 4.
Parameters of models for cross-receptors prediction (R2).
|
ACKNOWLEDGEMENTS
The work was carried out within the framework of the state task for 2018 (topic number 0090-2017-0020). The software tuning on hybrid cluster was supported by the Russian Foundation for Basic Research (project 18-29-03100).
REFERENCES
- Eglen, R. M. (2006). Muscarinic receptor subtypes in neuronal and non?neuronal cholinergic function. Autonomic and Autacoid Pharmacology, 26(3), 219-233. DOI
- Langmead, C. J., Watson, J., & Reavill, C. (2008). Muscarinic acetylcholine receptors as CNS drug targets. Pharmacology & therapeutics, 117(2), 232-243. DOI
- Freedman, S. B., Dawson, G. R., Iversen, L. L., Baker, R., & Hargreaves, R. J. (1993). The design of novel muscarinic partial agonists that have functional selectivity in pharmacological preparations in vitro and reduced side-effect profile in vivo. Life sciences, 52(5-6), 489-495. DOI
- Mikurova, A. V., Rybina, A. V., & Skvortsov, V. S. (2016). Prediction of selective inhibition of neuraminidase from various influenza virus strains by potential inhibitors. Biomeditsinskaya khimiya, 62(6), 691-703. DOI
- Thal, D. M., Sun, B., Feng, D., Nawaratne, V., Leach, K., Felder, C. C., ... & Kobilka, T. S. (2016). Crystal structures of the M1 and M4 muscarinic acetylcholine receptors. Nature, 531(7594), 335.
- Haga, K., Kruse, A. C., Asada, H., Yurugi-Kobayashi, T., Shiroishi, M., Zhang, C., ... & Kobayashi, T. (2012). Structure of the human M2 muscarinic acetylcholine receptor bound to an antagonist. Nature, 482(7386), 547.
- Kruse, A. C., Hu, J., Pan, A. C., Arlow, D. H., Rosenbaum, D. M., Rosemond, E., ... & Shaw, D. E. (2012). Structure and dynamics of the M3 muscarinic acetylcholine receptor. Nature, 482(7386), 552.
- Blundell, T., Carney, D., Gardner, S., Hayes, F., Howlin, B., Hubbard, T., ... & Sutcliffe, M. (1988). Knowledge?based protein modelling and design. European Journal of Biochemistry, 172(3), 513-520. DOI
- Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., ... & Bourne, P. E. (2000). The protein data bank. Nucleic acids research, 28(1), 235-242. DOI
- SYBYL-X 2.1. Certara, Princeton, NJ, USA.
- Halgren, T. A. (1996). Merck molecular force field. II. MMFF94 van der Waals and electrostatic parameters for intermolecular interactions. Journal of Computational Chemistry, 17(5/6), 520-552. DOI
- Scapecchi, S., Marucci, G., Matucci, R., Angeli, P., Bellucci, C., Buccioni, M., ... & Teodori, E. (2001). Structure–activity relationships in 2, 2-diphenyl-2-ethylthioacetic acid esters: unexpected agonistic activity in a series of muscarinic antagonists. Bioorganic & medicinal chemistry, 9(5), 1165-1174. DOI
- Alexander, S. P., Christopoulos, A., Davenport, A. P., Kelly, E., Marrion, N. V., Peters, J. A., ... & Southan, C. (2017). The Concise Guide to PHARMACOLOGY 2017/18: G protein-coupled receptors. British journal of pharmacology, 174(S1). DOI
- Dei, S., Bellucci, C., Buccioni, M., Ferraroni, M., Guandalini, L., Manetti, D., ... & Romanelli, M. N. (2007). Synthesis, affinity profile, and functional activity of muscarinic antagonists with a 1-methyl-2-(2, 2-alkylaryl-1, 3-oxathiolan-5-yl) pyrrolidine structure. Journal of medicinal chemistry, 50(6), 1409-1413. DOI
- Kuntz, I. D., Blaney, J. M., Oatley, S. J., Langridge, R., & Ferrin, T. E. (1982). A geometric approach to macromolecule-ligand interactions. Journal of molecular biology, 161(2), 269-288. DOI
- Case, D. A., Cheatham, T. E., Darden, T., Gohlke, H., Luo, R., Merz, K. M., ... & Woods, R. J. (2005). The Amber biomolecular simulation programs. Journal of computational chemistry, 26(16), 1668-1688. DOI
- Massova, I., & Kollman, P. A. (2000). Combined molecular mechanical and continuum solvent approach (MM-PBSA/GBSA) to predict ligand binding. Perspectives in drug discovery and design, 18(1), 113-135. DOI
- Federal Research Center Computer Science and Control of Russian Academy of Sciences [Electronic resource]: site. - Moscow: FRC CS RAS.- URL: http://hhpcc.frccsc.ru (application date: 09/12/2018)