Improvement of the Exon Method for Rapid Synthesis of cDNA of the Rat Renalase Gene
Institute of Biomedical Chemistry, Pogodinskaya Street, 10, Moscow 119121, Russia; *e-mail: valfed38@yandex.ru
Keywords:rat renalase gene; exon; exon assembly; PCR; gene expression
DOI:10.18097/BMCRM00201
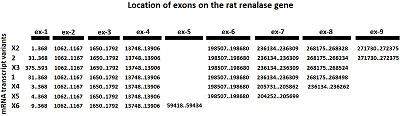
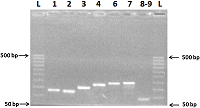
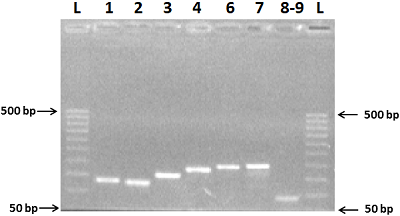
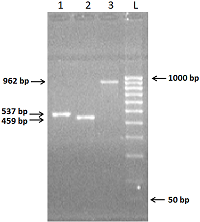
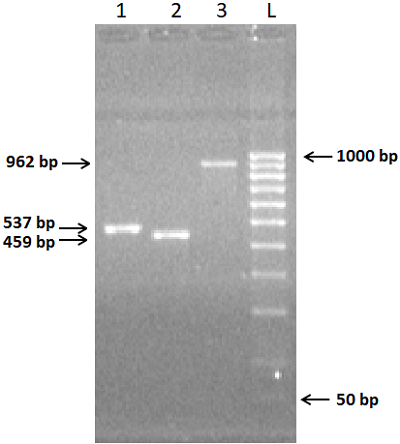
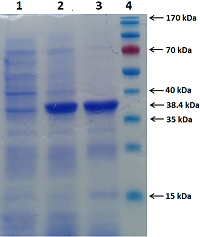
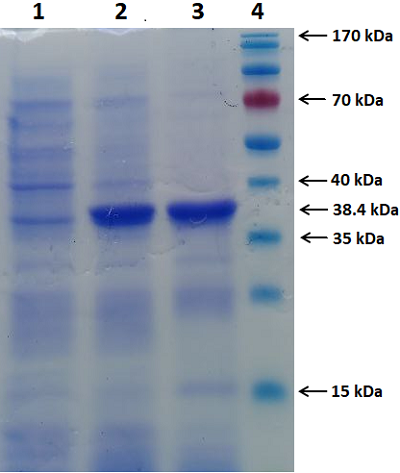
We have improved our previously developed method of exon cloning of cDNA of eukaryotic genes to obtain the rat renalase gene cDNA. In contrast to the previously used step-by-step pairwise assembly of exons, in this work the procedure of full-length cDNA preparation was shortened due to simultaneous assembly of four neighboring exons at once (exons 1-4 and exons 6-9 of the rat renalase gene). The two obtained sequences (exons 1-4 and 6-9) were combined into a full-length cDNA of the rat renalase gene. The cDNA synthesized in this way was cloned into the prokaryotic vector pET-28a(+), which was then expressed in E. coli cells. The correctness of this approach was confirmed by sequencing resultant cDNA sequencing, which showed full (100%) identity with the nucleotide sequence available in the GenBank database (accession code: GenBankNM_001014167).

|
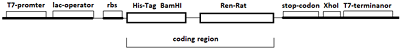
Figure 2.
The scheme of rat renalase gene cloning into the plasmid vector pET-28a(+) to obtain the vector pET-RenRatV2.
|
|
CLOSE

|
Table 1.
Primers used for PCR amplification to obtain exons (ex-1, ex-2, ex-3, ex-4, ex-6, ex-7, and ex-8+ex-9) of the mRNA transcription variant-2 of the rat renalase gene (Rattus norvegicus).
|
FUNDING
The work was performed within the framework of the Program for Basic Research in the Russian Federation for a long-term period (2021-2030) (№ 122030100170-5)
REFERENCES
- An, X., Lu, J., Huang, J.D., Zhang, X., Chen, J., Zhou, Y., Tong, Y. (2007) Rapid assembly of multiple-exon cDNA directly from genomic DNA. PLoSOne, 2, e1179. DOI
- Davies W.L., Carvalho L.S., Hunt D.M. (2007) SPLICE: A technique for generation in vitro spliced coding sequeces from genomic DNA. BioTechniques, 43, 785–789. DOI
- Jayakumar, A., Huang, W.-Y., Raetz, B., Chirala, S.S., Wakil, S.J. (1996) Cloning and expression of the multifunctional human fatty acid synthase and its subdomains in Escherichia coli. Proc. Natl. Acad. Sci. USA, 93, 14509–14514. DOI
- Booth, P.M., Buchman, G.W., Rashtchian, A. (1994) Assembly and cloning of coding sequences for neurotrophic factors directly from genomic DNA using polymerase chain reaction and uracil DNA glycosylase. Gene, 146, 303–308. DOI
- Fedchenko, V.I., Kaloshin, A.A., Mezhevikina, L.M., Buneeva, O.A., Medvedev, A.E. (2013) Construction of the Coding Sequence of the Transcription Variant 2 of the Human Renalase Gene and Its Expression in the Prokaryotic System.Int. J. Mol. Sci. v.14, 12764-12779; DOI
- Fedchenko, V.I., Kaloshin, A.A. (2019) A simplified method for obtaining cDNA of low-copy and silent eukaryotic genes using human renalase as an the example. Biomedical Chemistry: Research and /Content/2023/201/, 2(2), 1-7. DOI
- Fedchenko, V.I., Kaloshin, A.A., Kaloshina, S.A., Medvedev, A.E. (2021). Expression and Isolation of N-Terminal Truncated Human Recombinant Renalase in Prokaryotic Cells. Biomedical Chemistry: Research and /Content/2023/201/, 4(3), e00158. DOI
- Tersus Plus PCR kit. Retrived June 01, 2023 from https://evrogen.ru/kit-user-manuals/Tersus_PLUS_PK221.pdf