Strategy for Experimental Studies of Target Protein Interactomics
Institute of Biomedical Chemistry, 10 Pogodinskaya str., Moscow, 119121 Russia; *e-mail: gnedenko.oksana@gmail.com
Keywords: protein-protein interactions; affine selection; protein subinteractome; complexation; surface plasmon resonance; platform
DOI:10.18097/BMCRM00224
It is known that intermolecular interactions of proteins and peptides play a critical role in life processes. Such interactions can be either directly related to the implementation of various functions or play the role of a regulator. Currently, there is no doubt that the majority of proteins function as part of various molecular complexes, the formation of which occurs due to protein-protein interactions (PPIs), the totality of which can be defined as the “protein interactome”. Protein subinteractome studies are critical for studying the functions and regulatory mechanisms of unknown or poorly annotated proteins, understanding the architecture of intracellular molecular machines, and the design of PPI modulators. Previously, we used combinations of experimental approaches, as well as analytical and preparative methods, to study the subinteractomes of functionally different cellular proteins, which allowed us to identify the protein subinteractomes of several clinically significant human proteins. The purpose of this work was to conceptualize the principles of the experimental platform we developed for studying protein subinteractomes and to describe its features in detail.
|
CLOSE

|
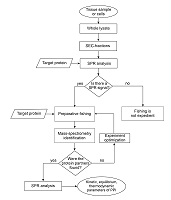
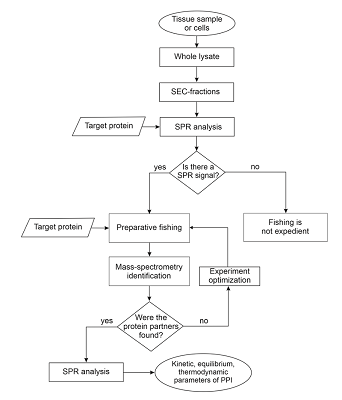
Table 1.
Structure of the experimental platform for studying the subinteractomes of target proteins.
|
FUNDING
The research was supported by the Russian Science Foundation grant No. 23-15-00149.
REFERENCES
- Bludau, I., Aebersold, R. (2020) Proteomic and interactomic insights into the molecular basis of cell functional diversity. Nat. Rev. Mol. Cell Biol., 21(6), 327–340. DOI
- Luck, K., Sheynkman, G.M., Zhang, I., Vidal, M. (2017) Proteome-scale human interactomics. Trends Biochem. Sci., 42(5), 342–354. DOI
- Ershov, P.V., Mezentsev, Y.V., Kopylov, A.T., Yablokov, E.O., Svirid, A.V., Lushchyk, A.Y., Kaluzhskiy, L.A., Gilep, A.A., Usanov, S.A., Medvedev, A.E., Ivanov, A.S. (2019) Affinity isolation and mass spectrometry identification of prostacyclin synthase (PTGIS) subinteractome. Biology (Basel), 8(2), 49. DOI
- Trisciuzzi, D., Villoutreix, B.O., Siragusa, L., Baroni, M., Cruciani, G., Nicolotti, O. (2023) Targeting protein-protein interactions with low molecular weight and short peptide modulators: Insights on disease pathways and starting points for drug discovery. Expert. Opin. Drug. Discov., 18(7), 737–752. DOI
- Ershov, P.V., Yablokov, E., Zgoda, V., Mezentsev, Y., Gnedenko, O., Kaluzhskiy, L., Svirid, A., Gilep, A., Usanov, S.A., Ivanov, A. (2021) A new insight into subinteractomes of functional antagonists: thromboxane (CYP5A1) and prostacyclin (CYP8A1) synthases. Cell Biol. Int., 45(6), 1175–1182. DOI
- Ershov, P., Mezentsev, Y., Gilep, A., Usanov, S., Buneeva, O., Medvedev, A., Ivanov, A. (2017) Isatin-induced increase in the affinity of human ferrochelatase and adrenodoxin reductase interaction. Protein Science, 26(12), 2458–2462. DOI
- Nakamura, K., Yamada, Y., Araki, K. (1986) [Nursing of patients with thoracic injuries]. Kango Gijutsu, 32(9), 1146–1151.
- Carregari, V.C. (2022) Protein extraction and sample preparation methods for shotgun proteomics with central nervous system cells and brain tissue. Adv. Exp. Med. Biol., 1382, 1–15. DOI
- Dapic, I., Baljeu-Neuman, L., Uwugiaren, N., Kers, J., Goodlett, D.R., Corthals, G.L. (2019) Proteome analysis of tissues by mass spectrometry. Mass Spectrom. Rev., 38(4–5), 403–441. DOI
- Cheerathodi, M.R., Meckes, D.G. (2020) BioID combined with mass spectrometry to study herpesvirus protein-protein interaction networks. Methods Mol. Biol., 2060, 327–341. DOI
- Ershov, P.V., Mezentsev, Y.V., Yablokov, E.O., Kaluzhskiy, L.A., Vakhrushev, I.V., Gnedenko, O.V., Florinskaya, A.V., Gilep, A.A., Usanov, S.A., Yarygin, K.N., Ivanov, A.S. (2019) A method of lysate preparation to improve the isolation efficiency of protein partners for target proteins encoded by the genes of human chromosome 18. Biomedical Chemistry: Research and Methods, 2(1), e00090. DOI
- Florinskaya, A., Ershov, P., Mezentsev, Y., Kaluzhskiy, L., Yablokov, E., Medvedev, A., Ivanov, A. (2018) SPR biosensors in direct molecular fishing: Implications for protein interactomics. Sensors (Switzerland), 18(5), 1616. DOI
- Florinskaya, A.V., Ershov, P.V., Mezentsev, Y.V., Kaluzhskiy, L.A., Yablokov, E.O., Buneeva, O.A., Zgoda, V.G., Medvedev, A.E., Ivanov, A.S. (2018) The analysis of participation of individual proteins in the protein interactome formation. Biomeditsinskaya Khimiya, 64(2), 169–174. DOI
- Kaiser, P., Meierhofer, D.,Wang, X., Huang, L. (2008) Tandem affinity purification combined with mass spectrometry to identify components of protein complexes. Methods Mol. Biol., 439, 309–326. DOI
- Liu, G., Fu, T., Han, Y., Hu, S., Zhang, X., Zheng, M., Hao, P., Pan, L., Kang, J. (2020) Probing protein-protein interactions with label-free mass spectrometry quantification in combination with affinity purification by spin-tip affinity columns. Anal. Chem., 92(5), 3913–3922. DOI
- Pardo, M., Choudhary, J.S. (2012) Assignment of protein interactions from affinity purification/mass spectrometry data. J. Proteome Res., 11(3), 1462-1474. DOI
- Jia, Y., Chen, S., Wang, Q., Li, J. (2024) Recent progress in biosensor regeneration techniques. Nanoscale, 16(6), 2834–2846. DOI
- Food and Drug Administration,HHS (2019) Immunogenicity testing of therapeutic protein products-developing and validating assays for anti-drug antibody detection; Guidance for industry. Docket No. FDA-2009-D-0539,.
- Thoren, K.L., Pasi, B., Delgado, J.C., Wu, A.H.B., Lynch, K.L. (2018) Quantitation of infliximab and detection of antidrug antibodies in serum by use of surface plasmon resonance. J. Appl. Lab. Med., 2(5), 725–736. DOI
- Li, Y., Franklin, S., Zhang, M.J., Vondriska, T.M. (2011) Highly efficient purification of protein complexes from mammalian cells using a novel streptavidin-binding peptide and hexahistidine tandem tag system: Application to Bruton's tyrosine kinase. Protein Science, 20(1), 140–149. DOI
- Bludau, I., Heusel, M., Frank, M., Rosenberger, G., Hafen, R., Banaei-Esfahani, A., van Drogen, A., Collins, B.C., Gstaiger, M., Aebersold, R. (2020) Complex-centric proteome profiling by SEC-SWATH-MS for the parallel detection of hundreds of protein complexes. Nature Protocols, 15(8), 2341–2386. DOI
- DasGupta, B.R., Boroff, D.A. (1968) Separation of toxin and hemagglutinin from crystalline toxin of Clostridium botulinum typeAby anion exchange chromatography and determination of their dimensions by gel filtration. J. Biol. Chem., 243(5), 1065–1072.
- Gilbert, M., Schulze, W.X. (2019) Global identification of protein complexes within the membrane proteome of arabidopsis roots using a SEC-MS approach. J. Proteome Res., 18(1), 107–119. DOI
- Fossati, A., Frommelt, F., Uliana, F., Martelli, C., Vizovisek,M., Gillet, L., Collins, B., Gstaiger, M., Aebersold, R. (2021) System-wide profiling of protein complexes via size exclusion chromatography-mass spectrometry (SEC-MS). Methods Mol. Biol., 2259, 269–294. DOI
- Wittig, I., Malacarne, P.F. (2021) Complexome profiling: Assembly and remodeling of protein complexes. Int. J. Mol. Sci., 22(15), 7809. DOI
- Heusel, M., Bludau, I., Rosenberger, G., Hafen, R., Frank, M., Banaei-Esfahani, A., van Drogen, A., Collins, B.C., Gstaiger, M., Aebersold, R. (2019) Complex-centric proteome profiling by SEC-SWATH-MS. Mol. Syst. Biol., 15(1), e8438. DOI
- Ershov, P.V., Mezentsev, Y.V., Yablokov, E.O., Kaluzhsky, L.A., Florinskaya, A.V., Buneeva, O.A., Medvedev, A.E., Ivanov, A.S. (2018) Effect of bioregulator isatin on protein-protein interactions involving isatin-binding proteins. Russ. J. Bioorg. Chem., 44(2), 193–198. DOI
- Yablokov, E.O., Sushko, T.A., Ershov, P.V., Florinskaya, A.V., Gnedenko, O.V., Shkel, T.V., Grabovec, I.P., Strushkevich, N.V., Kaluzhskiy, L.A., Usanov, S.A., Gilep, A.A., Ivanov, A.S. (2019) A large-scale comparative analysis of affinity, thermodynamics and functional characteristics of interactions of twelve cytochrome P450 isoforms and their redox partners. Biochimie, 162, 156–166. DOI
- Campbell, L., Simpson, D., Ramasamy, K., Sadler, R. (2021) Using quantitative immunoprecipitation mass spectrometry (QIP-MS) to identify low level monoclonal proteins. Clin. Biochem., 95, 81–83. DOI
- Jensen, P., Patel, B., Smith, S., Sabnis, R., Kaboord, B. (2021) Improved immunoprecipitation to mass spectrometry method for the enrichment of low-abundant protein targets. Methods Mol. Biol., 2261, 229–246. DOI
- Jerabek-Willemsen, M., Wienken, C.J., Braun, D., Baaske, P., Duhr, S. (2011) Molecular interaction studies using microscale thermophoresis. Assay Drug Dev. Technol., 9(4), 342–353. DOI
- Nair, M.P., Teo, A.J.T., Li, K.H.H. (2021) Acoustic biosensors and microfluidic devices in the decennium: Principles and applications. Micromachines (Basel), 13(1), 24. DOI
- Basavarajappa, H.D., Sulaiman, R.S., Qi, X., Shetty, T., Sheik Pran Babu, S., Sishtla, K.L., Lee, B., Quigley, J., Alkhairy, S., Briggs, C.M., Gupta, K., Tang, B., Shadmand, M., Grant, M.B., Boulton, M.E., Seo, S.-Y., Corson, T.W. (2017) Ferrochelatase is a therapeutic target for ocular neovascularization. EMBO Mol. Med., 9(6), 786–801. DOI
- Sishtla, K., Lambert-Cheatham, N., Lee, B., Han, D.H., Park, J., Sardar Pasha, S.P.B., Lee, S., Kwon, S., Muniyandi, A., Park, B., Odell, N., Waller, S., Park, I.Y., Lee, S.J., Seo, S.-Y., Corson, T.W. (2022) Small-molecule inhibitors of ferrochelatase are antiangiogenic agents. Cell Chem. Biol., 29(6), 1010–1023.e14. DOI
- Burden, A.E., Wu, C., Dailey, T.A., Busch, J.L., Dhawan, I.K., Rose, J.P., Wang, B., Dailey, H.A. (1999) Human ferrochelatase: Crystallization, characterization of the [2Fe-2S] cluster and determination that the enzyme is a homodimer. Biochim. Biophys. Acta, 1435(1–2), 191–197. DOI
- Obi, C.D., Dailey, H.A., Jami-Alahmadi, Y.,Wohlschlegel, J.A., Medlock, A.E. (2023) Proteomic analysis of ferrochelatase interactome in erythroid and non-erythroid cells. Life (Basel), 13(2), 577. DOI
- Ivanov, A.S., Ershov, P.V., Molnar, A.A., Mezentsev, Yu.V., Kaluzhskiy, L.A., Yablokov, E.O., Florinskaya, A.V., Gnedenko, O.V., Medvedev, A.E., Kozin, S.A., Mitkevich, V.A., Makarov, A.A., Gilep, A.A., Luschik, A.Ya., Gaidukevich, I.V., Usanov, S.A. (2016) Direct molecular fishing in molecular partners investigation in protein-protein and protein-peptide interactions. Russ. J. Bioorg. Chem., 42(1), 14–21. DOI
- Sheftel, A.D., Stehling, O., Pierik, A.J., Elsasser, H.-P., Muhlenhoff, U., Webert, H., Hobler, A., Hannemann, F., Bernhardt, R., Lill, R. (2010) Humans possess two mitochondrial ferredoxins, Fdx1 and Fdx2, with distinct roles in steroidogenesis, heme, and Fe/S cluster biosynthesis. Proc. Natl. Acad. Sci. USA, 107(26), 11775–11780. DOI
- Ershov, P.V., Veselovsky, A.V., Mezentsev, Y.V., Yablokov, E.O., Kaluzhskiy, L.A., Tumilovich, A.M., Kavaleuski, A.A., Gilep, A.A., Moskovkina, T.V., Medvedev, A.E., Ivanov, A.S. (2020) Mechanism of the affinity-enhancing effect of isatin on human ferrochelatase and adrenodoxin reductase complex formation: Implication for protein interactome regulation. Int. J. Mol. Sci., 21(20), 7605. DOI
- Biringer, R.G. (2020) The enzymology of the human prostanoid pathway. Mol. Biol. Rep., 47(6), 4569–4586. DOI
- Beccacece, L., Abondio, P., Bini, C., Pelotti, S., Luiselli, D. (2023) The link between prostanoids and cardiovascular diseases. Int. J. Mol. Sci., 24(4), 4193. DOI
- Svirid, A.V., Ershov, P.V., Yablokov, E.O., Kaluzhskiy, L.A., Mezentsev, Y.V., Florinskaya, A.V., Sushko, T.A., Strushkevich, N.V., Gilep, A.A., Usanov, S.A., Medvedev, A.E., Ivanov, A.S. (2017) Direct molecular fishing of new protein partners for human thromboxane synthase. Acta Naturae, 9(4), 92–100. DOI