MetaPASS 2024: VISUALIZATION OF BIOLOGICAL ACTIVITY SPECTRA OF ORGANIC COMPOUNDS TAKING INTO ACCOUNT THEIR BIOTRANSFORMATION
1Institute of Biomedical Chemistry, 10 bldg. 8, Pogodinskaya str., Moscow, 119121; Russia *e-mail: rudik_anastassia@mail.ru
2Pirogov Russian National Research Medical University, 1, Ostrovitianov str., Moscow, 117997, Russia
Keywords: drug-like compounds; biological activity spectrum; xenobiotic metabolism; treemap
DOI:10.18097/BMCRM00243
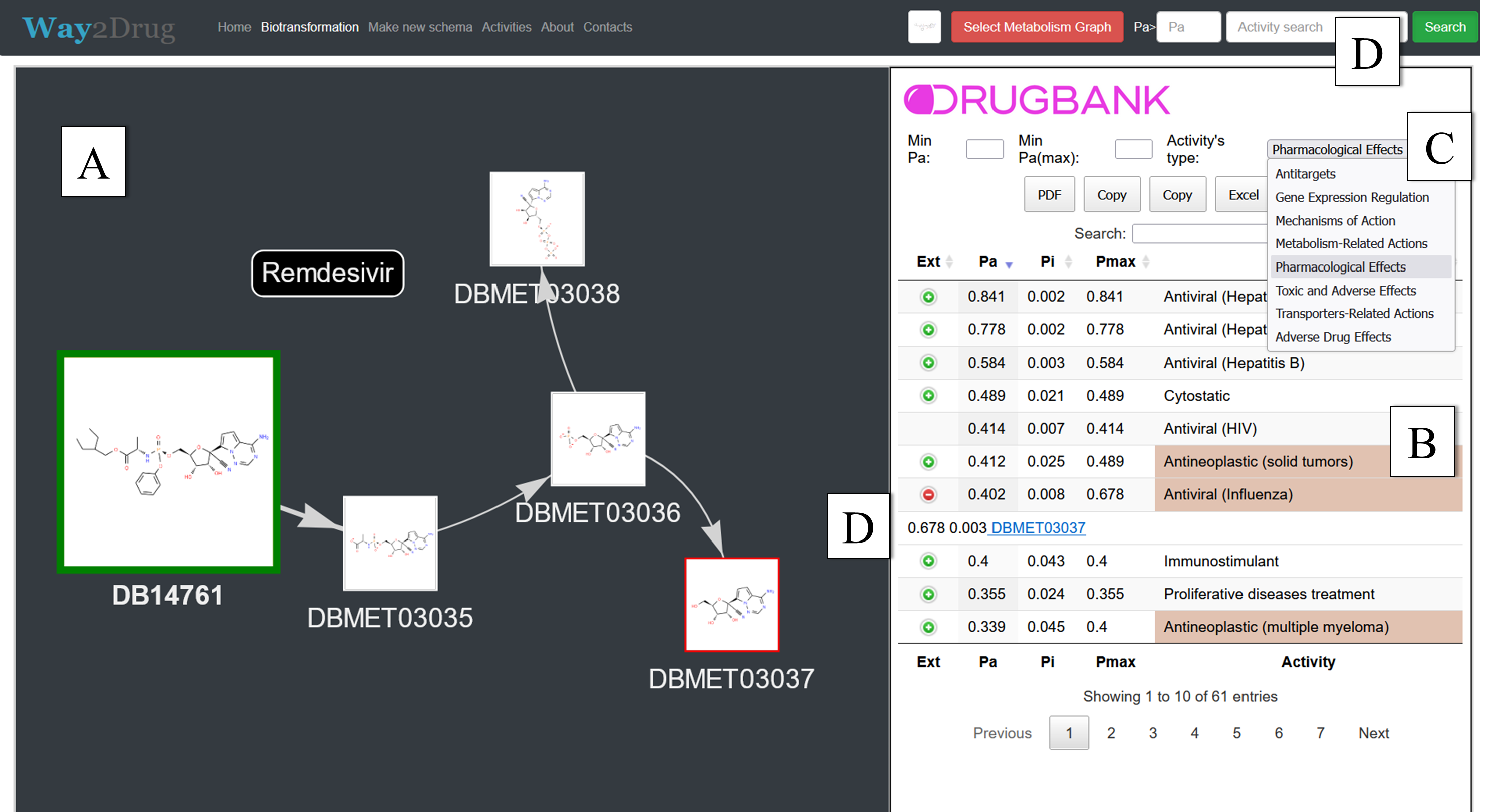
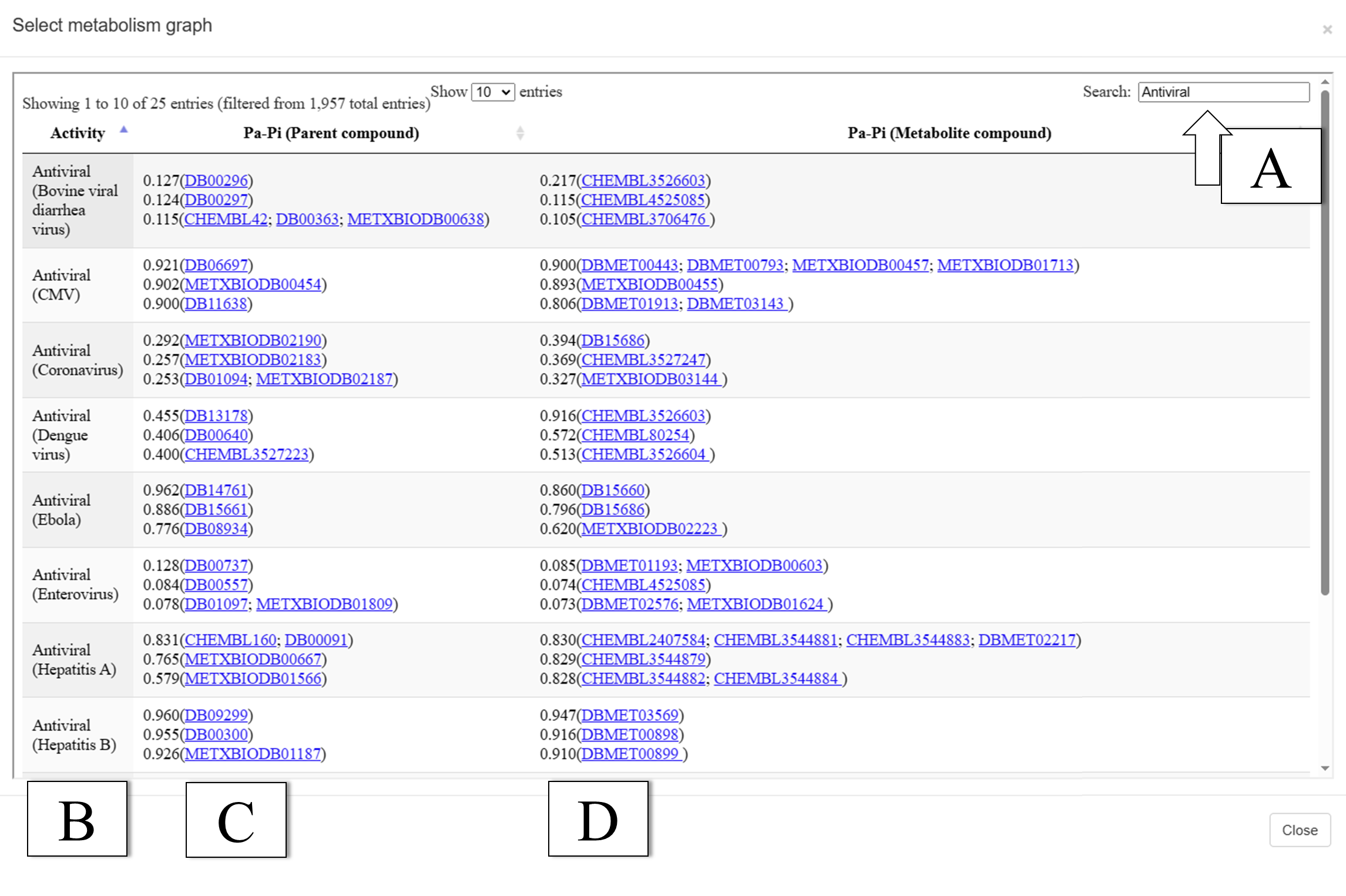
In the human body, pharmacological substances undergo biotransformation, therefore, during drugs development, it is necessary to take into account the biological activity spectra of their metabolites. Previously, we created the MetaPASS web application to analyze the probable spectra of biological activity of drug-like organic compounds taking into account their metabolism. Here we describe a new version of MetaPASS 2024 (https://www.way2drug.com/metapass), containing increased number of known metabolic pathways, and added procedures for searching structural similarity based on MNA and QNA descriptors and searching for compounds with the highest probability estimate for target biological activity; we have also implemented representation of the spectrum of biological activity in the form of treemaps.
|
CLOSE

|
Table 1.
Data replenishment in MetaPASS.
|
|
CLOSE

|
Table 2.
Color coding for activity categories.
|
|
CLOSE

|
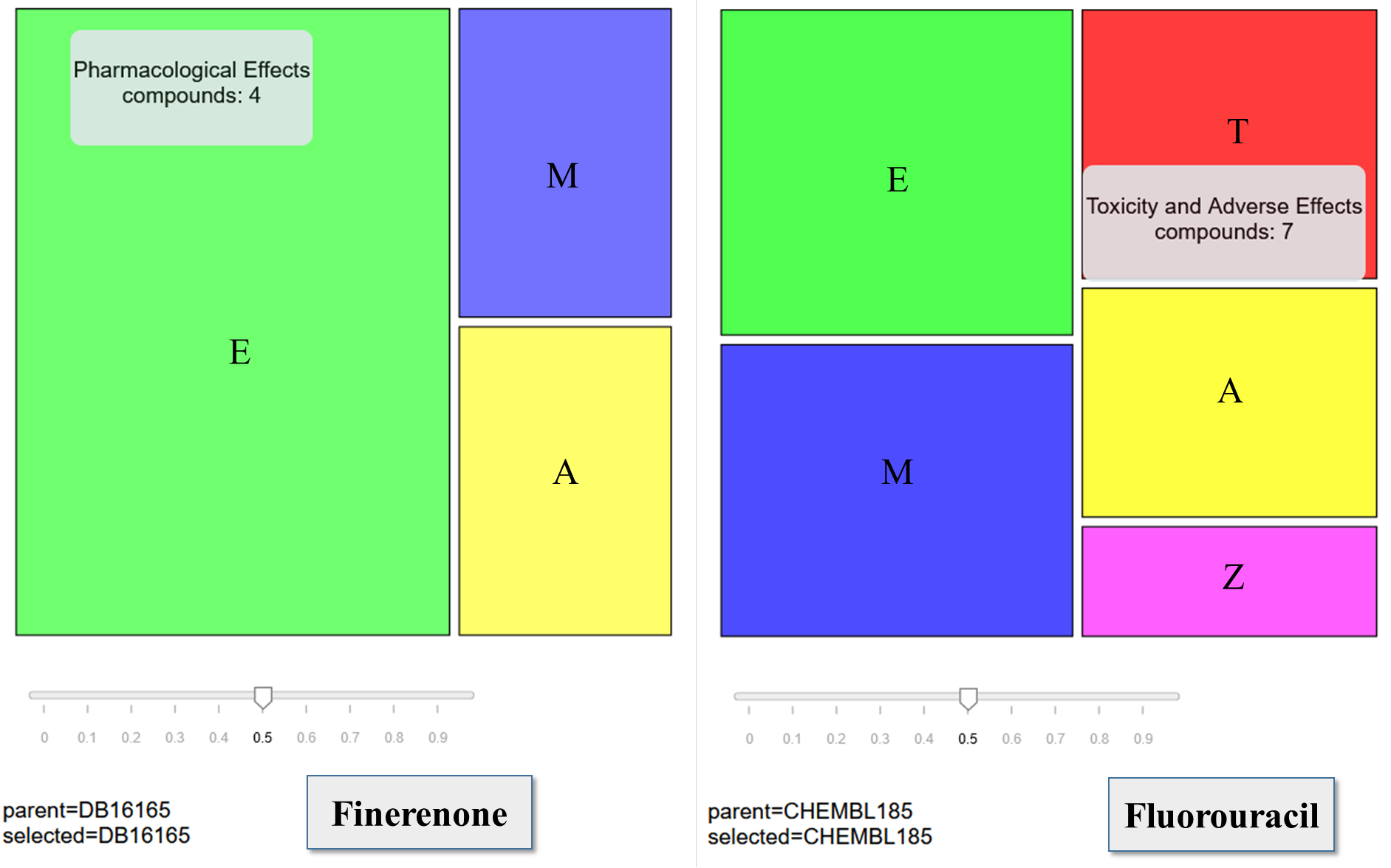
Table 3.
The biological activity spectrum prediction in the « Pharmacological Effects » category at the cutoff value of Pa- Pi > 0.5 for Finerenone. DBMET03393 and DBMET03398 are Finerenone’s metabolites
|
|
CLOSE

|
Table 4.
The study was carried out within the Programme for Basic Scientific Research in the Russian Federation for the Long-Term Period (2021 – 2030 years) (№ 122030100170-5).
|
FUNDING
The research was supported by the Russian Science Foundation grant No. 23-15-00149.
REFERENCES
- DiMasi, J.A., Grabowski, H.G., Hansen, R.W. (2016) Innovation in thepharmaceutical industry: New estimates of R&D costs. J. Health Econ. 47,20–33. DOI
- Martinez-Mayorga, K., Madariaga-Mazon, A., Medina-Franco, J.L.,Maggiora, G. (2020) The impact of chemoinformatics on drug discovery in thepharmaceutical industry. Expert Opin. Drug Discov. 15(3), 293–306. DOI
- Muratov, E.N., Bajorath, J., Sheridan, R.P., Tetko, I. V, Filimonov, D.,Poroikov, V., Oprea, T.I., Baskin, I.I., Varnek, A., Roitberg, A., Isayev, O.,Curtarolo, S., Fourches, D., Cohen, Y., Aspuru-Guzik, A., Winkler, D.A.,Agrafiotis, D., Cherkasov, A., Tropsha, A. (2020) QSAR without borders. Chem.Soc. Rev. 49(11), 3525–3564. DOI
- Pushpakom, S., Iorio, F, Eyer,s P.A., Escott, K.J., Hopper, S., Wells, A., Doig,A., Guilliams, T., Latimer, J., McNamee, C., Norris, A., Sanseau, P., Cavalla,D., Pirmohamed, M. (2019) Drug repurposing: progress, challenges andrecommendations. Nat. Rev. Drug Discov. 18, 41–58. DOI
- Eno, M.R., Cameron, M.D. (2015) Gauging reactive metabolites in druginducedtoxicity. Curr. Med. Chem. 22 (4), 465–489. DOI
- Rudik, A.V., Dmitriev, A.V., Lagunin, A.A., Filimonov, D.A., Poroikov, V.V.(2021) MetaPASS: A Web Application for Analyzing the Biological ActivitySpectrum of Organic Compounds Taking into Account their Biotransformation.Mol. Inform. 40(4), e2000231.
- Rudik, A.V., Dmitriev, A.V., Lagunin, A.A., Filimonov, D.A., Poroikov, V.V.(2023) MetaTox 2.0: Estimating the Biological Activity Spectra of Drug-likeCompounds Taking into Account Probable Biotransformations. ACS Omega.8(48), 45774-45778. DOI
- Filimonov, D.A., Druzhilovskiy, D.S., Lagunin, A.A., Gloriozova, T.A.,Rudik, A.V, Dmitriev A.V, Pogodin P.V., Poroikov, V.V. (2018) ComputeraidedPrediction of Biological Activity Spectra for Chemical Compounds:Opportunities and Limitations, Biomedical Chemistry: Research and Methods,1(1), e00004. DOI
- MarvinJS-demo. Chemaxon. Retrieved May 17, 2025, from: https://marvinjsdemo.chemaxon.com/latest/
- Filimonov, D.A., Zakharov, A.V., Lagunin, A.A., Poroikov, V.V. (2009)QNA-based ‘Star Track’ QSAR approach. SAR QSAR Environ Res. 20(7-8),679-709. DOI
- Mendez, D., Gaulton, A., Bento, A.P., Chambers, J., De Veij, M., Félix, E.,Magariños, M.P., Mosquera, J.F., Mutowo, P., Nowotka, M., Gordillo-Marañón,M., Hunter, F., Junco, L., Mugumbate, G., Rodriguez-Lopez, M., Atkinson, F.,Bosc, N., Radoux, C.J., Segura-Cabrera, A., Hersey, A., Leach, A.R. (2018)ChEMBL : towards direct deposition of bioassay data. Nucleic Acids Res.,47(1), 930-940. DOI
- Wishart, D.S., Feunang, Y.D., Guo, A.C., Lo E.J., Marcu, A., Grant, J.R.,Sajed, T., Johnson, D., Li C., Sayeeda, Z., Assempour, N., Iynkkaran, I., Liu Y.,Maciejewski, A., Gale, N., Wilson, A., Chin, L., Cummings, R., Le, D., Pon, A.,Knox, C., Wilson, M. (2018), DrugBank 5.0 : a major update to the DrugBankdatabase for 2018. Nucleic Acids Res., 46 (1), 1074–1082.
- Djoumbou-Feunang, Y., Fiamoncini, J., Gil-de-la-Fuente, A., Greiner,R., Manach, C., Wishart, D.S. (2019) BioTransformer: a comprehensivecomputational tool for small molecule metabolism prediction and metaboliteidentification. J. Cheminform. 11, 2. DOI
- Dunlop, B.W., Nemeroff, C.B.(2007) The Role of Dopamine in thePathophysiology of Depression. Arch Gen Psychiatry, 64(3), 327–337. DOI
- Lagunin, A.A., Romanova, M.A., Zadorozhny, A.D., Kurilenko, N.S., Shilov,B.V, Pogodin, P.V., Ivanov, S.M., Filimonov, D.A., Poroikov, V. V. (2018)Comparison of Quantitative and Qualitative (Q)SAR Models Created for thePrediction of K(i) and IC(50) Values of Antitarget Inhibitors. Front. Pharmacol.9, 1136. DOI
- The JavaScript library for data visualization. Retrieved May 17, 2025, from:https://d3js.org/
- Johnson, B., Shneiderman, B. (1991) Tree-maps: a space-filling approach tothe visualization of hierarchical information structures. Proceeding Visualization’91, 284–291. DOI
- Lerma, E.V., Wilson, D.J. (2022) Finerenone: a mineralocorticoid receptorantagonist for the treatment of chronic kidney disease associated with type 2diabetes. Expert Rev. Clin. Pharmacol. 15, 501–513. DOI
- Latchman J., Guastella A., Tofthagen C. (2014) 5-Fluorouracil toxicity anddihydropyrimidine dehydrogenase enzyme: implications for practice. Clin JOncol Nurs. 18(5), 581-585. DOI
- Clementi, N., Scagnolari, C., D’Amore, A., Palombi, F., Criscuolo, E.,Frasca, F., Pierangeli, A., Mancini, N., Antonelli, G., Clementi, M., Carpaneto,A., Filippini, A. (2021) Naringenin is a powerful inhibitor of SARS-CoV-2infection in vitro. Pharmacol Res., 163, 105255. DOI
- Agrawal, P.K., Agrawal, C., Blunden, G. (2021) PharmacologicalSignificance of Hesperidin and Hesperetin, Two Citrus Flavonoids, asPromising Antiviral Compounds for Prophylaxis Against and CombatingCOVID-19, Nat. Prod. Commun. 16. DOI
- Bílek, R., Danzig, V., Grimmichová, T. (2022) Antiviral activity ofamiodarone in SARS-CoV-2 disease. Physiol. Res. 71, 869–875. DOI
- Gasm,i A., Mujawdiya, P.K., Lysiuk, R., Shanaida, M., Peana, M., GasmiBenahmed, A., Beley, N., Kovalska, N., Bjørklund, G. (2022) Quercetin in thePrevention and Treatment of Coronavirus Infections: A Focus on SARS-CoV-2.Pharmaceuticals (Basel). 15(9), 1049. DOI
- Roy, A.V., Chan, M., Banadyga, L., He, S., Zhu, W., Chrétien, M., Mbikay,M. (2024) Quercetin inhibits SARS-CoV-2 infection and prevents syncytiumformation by cells co-expressing the viral spike protein and human ACE2. Virol.J. 21, 29. DOI