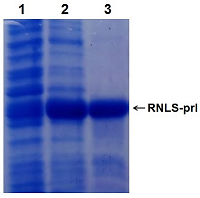
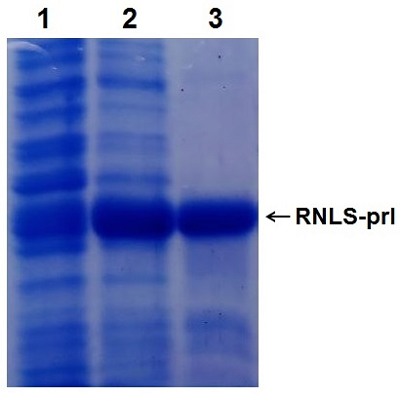
Construction and Expression of the Chimeric Human Renalase Gene Encoding the N-Terminal Signal Sequence of the Secretory Protein Prolactin
Institute of Biomedical Chemistry, 10 Pogodinskaya str., Moscow, 119121 Russia; *e-mail: valfed38@yandex.ru
Keywords:PCR; cloning; expression; renalase; prolactin; signal sequence
DOI:10.18097/BMCRM00175
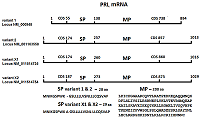
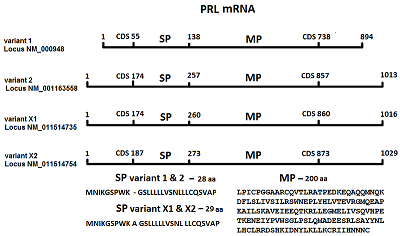
Renalase (RNLS) is a protein that performs various protective functions both inside and outside cells. Intracellular RNLS is a FAD-dependent oxidoreductase (EC 1.6.3.5). Extracellular RNLS lacking an N-terminal peptide does not interact with FAD and exhibits various protective effects on the cell through interaction with receptor proteins. The mechanisms and factors responsible for RNLS transport out of the cell are not fully understood. It is well known that the signal sequence plays a key role in the classical mechanism of protein transport outside cells. One of the approaches to study the secretion of RNLS from the cell can be the creation of chimeric forms of the protein with a modified N-terminal amino acid signal sequence. Bioinformatics analysis showed that the signal sequence of the prolactin gene (PRL), connected to the template sequence of the RNLS gene, gave the classic signal characteristic of secretory proteins. On this basis, this paper describes: (i) a method for constructing the human RNLS gene in which the N-terminal sequence encoded by the RNLS gene was replaced by the N-terminal sequence encoded by the PRL gene; (ii) expression of this chimeric genetic construct.
|
CLOSE

|
Table 1.
Prediction of the presence of the signal peptide (SP) in chimeric renalases.
|
|
CLOSE

|
Table 2.
Prediction of cellular localization of renalase with the N-terminal peptide from other proteins.
|
|
CLOSE

|
Table 3.
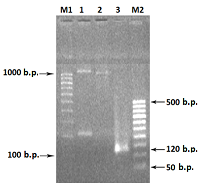
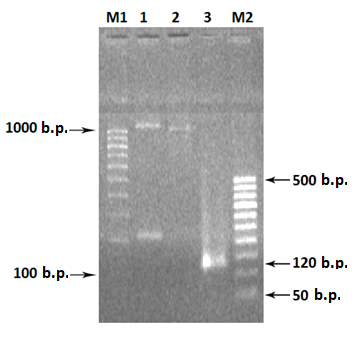
Primers used in PCR of the chimeric RNLS gene .
|
FUNDING
This work was supported by the Russian Foundation for Basic Research (project no. 20–015–00104).
REFERENCES
- Xu, J., Li, G., Wang, P., Velazquez, H., Yao, X., Li, Y., Wu, Y., Peixoto, A., Crowley, S., Desir, G.V. (2005) Renalase is a novel, soluble monoamine oxidase that regulates cardiac function and blood pressure. J. Clin. Invest., 115(5), 1275–1280. DOI
- Medvedev, A.E., Veselovsky, A.V., Fedchenko, V.I. (2010) Renalase, a new secretory enzyme responsible for selective degradation of catecholamines: achievements and unsolved problems. Biochemistry (Moscow), 75(8), 951-958. DOI
- Baroni, S., Milani, M., Pandini, V., Pavesi, G., Horner, D., Aliverti, A. (2013) Isrenalase a novel player in catecholaminergic signaling? The mystery of the catalytic activity of an intriguing new flavoenzyme. Curr. Pharm. Des., 19, 2540-2551. DOI
- Desir, G.V., Peixoto, A.J. (2014) Renalase in hypertension and kidney disease. Nephrol. Dial. Transplant., 29(1), 22-28. DOI
- Moran, G.R. (2016) The catalytic function of renalase: A decade of phantoms. BiochimBiophysActa, 1864(1), 177-186. DOI
- Moran, G.R., Hoag, M.R. (2017) The enzyme: Renalase. Arch. BiochemBiophys, 632, 66-76. DOI
- Milani, M., Ciriello, F., Baroni, S., Pandini V., Canevari, G., Bolognesi, M., Aliverti, A. (2011) FAD-binding site and NADP reactivity in human renalase: a new enzyme involved in blood pressure regulation. J. Mol. Biol., 411(2), 463-473. DOI
- Fedchenko, V.I,. Buneeva, O.A., Kopylov, A.T., Veselovsky, A.V., Zgoda, V.G., Medvedev, A.E. (2015) Human urinary renalase lacks the N-terminal signal peptide crucialfor accommodation of its FAD cofactor. International Journal of Biological Macromolecules, 78, 347–353. DOI
- Wang, Y., Safirstein, R., Velazquez, H., Guo, X.J., Hollander, L., Chang, J., Chen, T.M., Mu, J.J., Desir, G.V. (2017) Extracellular renalase protects cells and organs by outside-in signalling. J Cell Mol Med., 21(7), 1260-1265. DOI
- Kolodecik, T.R., Reed, A.M., Date, K., Shugrue, C.A., Patel, V., Chung, S.L.,
- Desir, G.V., Gorelick, F.S. (2017) The serum protein renalase reduces injury in experimentalpancreatitis. J. Biol. Chem., 292(51), 21047–21059. DOI
- 11. Wang, L., Velazquez, H., Chang, J., Safirstein, R., Desir, G.V. (2015) Identification of a receptor for extracellular renalase. PLoS One, 10, e0122932. DOI
- Fedchenko, V., Kopylov, A., Kozlova, N., Buneeva, O., Kaloshin, A., Zgoda, V., Medvedev, A. (2016) Renalase Secreted by Human Kidney НЕК293Т Cells Lacks its N-Terminal Peptide: Implications for Putative Mechanisms of Renalase Action. Kidney Blood Press Res., 41, 593-603. DOI
- Fedchenko, V.I., Kaloshin, A.A., Kozlova, N.I., Kopylov, A.T., Medvedev, A.E.(2020) Construction of a chimeric human gene encoding renalase with a modified N-terminus. Biomedical Chemistry: Research and Methods, 3(3), e00137. DOI
- Fedchenko, V.I., Kaloshin, A.A. (2019) A simplified method for obtaining cDNA of low-copy and silent eukaryotic genes using human renalase as anexample. Biomedical Chemistry: Research and Methods. 2(2), e00101. DOI
- Fedchenko, V.I.,. Kaloshin, A.A., Mezhevikina, L.M., Buneeva, O., Medvedev A.E. (2013) Construction of the Coding Sequence of the Transcription Variant 2 of the Human RenalaseGene and Its Expression in the Prokaryotic System, Int. J. Mol. Sci. 14, (6), 12764-7779. DOI
- Käll, L., Krogh, A., Sonnhammer, E.L.L. (2007) Advantages of combined transmembrane topology and signal peptide prediction--the Phobius web server, Nucleic Acids Res., 35, W429-32. DOI
- Juan, J., Armenteros, A., Salvatore, M., Winther, O. (2019) Ol of Emanuelsson, Gunnar von Heijne, Arne Elofsson, and Henrik Nielsen. Detecting Sequence Signals in Targeting Peptides Using Deep Learning. Life Science Alliance, 2 (5), e201900429. DOI
- Juan, J., o Armenteros, A., Konstantinos, D., Tsirigos, C.K.S., Petersen, T.N., Winther, O., Brunak, S., Heijne, G., Nielsen, H. (2019) Signal P 5.0 improves signal peptide predictions using deep neural networks, Nature Biotechnology, 37, 420-423, DOI
- Bendtsen, J., Kiemer, D. L., Fausbøll, A,. Brunak, S. (2005) Non-classical protein secretion in bacteria.BMC Microbiology, 5, 58. DOI
- Bhasin, M., Raghava, G.P. (2004) ESLpred: SVM-based method for subcellular localization of eukaryotic proteins using dipeptide composition and PSI-BLAST. Nucleic Acids Res. 32(suppl_2), W414-419. DOI
- Shen, H.-B., Chou, K.-C. (2009) A top-down approach to enhance the power of predicting human protein subcellular localization: Hum-mPLoc 2.0, Analytical Biochemistry, 394, 269-274. DOI
- Pierleoni, A., Martelli, P.L., Fariselli, P., Casadio, R. (2006) BaCelLo: a balanced subcellular localization predictor. Bioinformatics, 15:22(14):e408-16. DOI
- Drozdetskiy, A., Cole, C., Procter, J., Barton, G.J. (2006) JPred4: a protein secondary structure prediction server. Nucleic Acids Res., 43(W1), W389–W394. DOI
- Buchan, D.W.A., Jones, D.T. (2019) The PSIPRED Protein Analysis Workbench: 20 years on. Nucleic Acids Res., 47(W1), W402-W407. DOI
- Garnier, J., Gibrat, J-F., Robson, B. (1996) GOR secondary structure prediction method version IV. Methods in Enzymology R.F. Doolittle Ed., 266, 540-553.
- Geourjon, C., Deleage, G. (1995) SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput. Appl. Biosci., 11(6), 681-684. DOI
- Lee, P.Y., Costumbrado, J., Hsu, C.Y., Kim, Y. H. (2012) Agarose Gel Electrophoresis for the Separation of DNA Fragments. J. Vis. Exp., 62, e3923, DOI
- Laemmli, U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature; 227, 680-685. DOI
- Kushner, S. R. (1978) An improved method for transformation of Escherichia coli with ColE1-derived plasmids. In: Genetic engineering (Boyer H.B. and Nicosia S., eds.), Elsevier, North-Holland, Amsterdam, p. 17
- Fedchenko, V.I., Kaloshin, A.A., Kaloshina, S.A., Medvedev, A.E. (2021) Expression and isolation of N-terminal trancated human recombinant renalase in prokariotic cells. Biomedical Chemistry: Research and Methods, 4(3), e00158. DOI
- Freeman, M. E., Kanyicska, B., Lerant, A., and Nagy, G. (2000) Prolactin: structure, function, and regulation of secretion. Physiol. Rev., 80, 1523–1631. DOI
- Fedchenko, V.I., Kaloshin, A.A. (2019) A simplified method for obtaining cDNA of low-copy and silent eukaryotic genes using human renalase as an example. Biomedical Chemistry: Research and Methods, 2(2), e00101. DOI