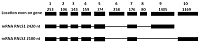
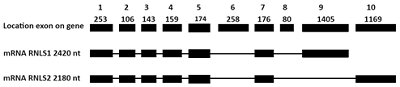
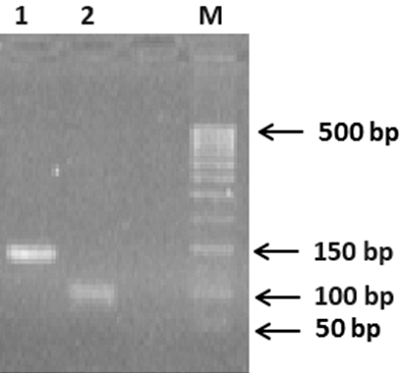
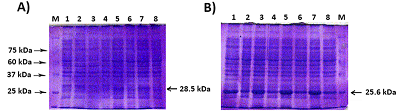
Generation of C-Terminal Sequences of Human Renalase-1 and Renalase-2 Encoded by Alternative Exons
Institute of Biomedical Chemistry, Pogodinskaya str., 10, Moscow 119121, Russia; *e-mail: valfed38@yandex.ru
Keywords:renalase (RNLS); alternative exons of the RNLS gene; RNLS1 and RNLS2 isoforms of renalase; cloning; expression; protein purification
DOI:10.18097/BMCRM00228
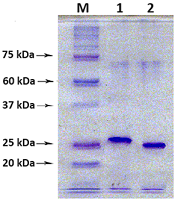
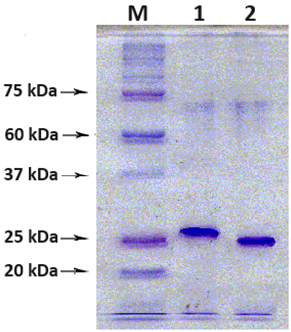
A method for generation of C-terminal amino acid sequences fused to dihydrofolate reductase (DHFR) and specific for RNLS1 and RNLS2 isoforms of renalase is described. It includes synthesis of nucleotide sequences of alternative exons of RNLS1-9ex and RNLS2-10ex, determining the differences in the primary structure of these proteins, their fusion with the coding sequence of DHFR and expression of these genetic constructs in cells of the E. coli Rosetta cells. Chromatographic purification on a column containing Ni Sepharose resulted in highly purified preparations of reombinant ReI-9ex and ReII-10ex proteins with an electrophoretic purity of about 95%.


|
Figure 3.
The scheme of RNLS1-9ex and RNLS2-10ex cloning into the pQE40 vector by BglII and PstI restriction sites. Explanations are given in the text.
|
|
CLOSE

|
Table 1.
Primers used for PCR-based amplification of RNLS-1-9ex и RNLS-2-10ex
|
FUNDING
The work was performed within the framework of the Program for Basic Research in the Russian Federation for a long-term period (2021-2030). (no. 122030100170-5).
REFERENCES
- Xu, J., Li, G., Wang, P., Velazquez, H., Yao, X., Li, Y., Wu, Y., Peixoto, A., Crowley, S., Desir, G.V. (2005) Renalase is a novel, soluble monoamine oxidase that regulates cardiac function and blood pressure. J. Clin. Invest., 115(5), 1275–1280. DOI
- Medvedev, A.E., Veselovsky, A.V., Fedchenko, V.I. (2010) Renalase, a new secretory enzyme responsible for selective degradation of catecholamines: achievements and unsolved problems. Biochemistry (Moscow), 75(8), 951-958. DOI
- Baroni, S., Milani, M., Pandini, V., Pavesi, G., Horner, D., Aliverti, A. (2013) Is renalase a novel player in catecholaminergic signaling? The mystery of the catalytic activity of an intriguing new flavoenzyme. Curr. Pharm. Des., 19, 2540-2551. DOI
- Desir, G.V., Peixoto, A.J. (2014) Renalase in hypertension and kidney disease. Nephrol. Dial. Transplant., 29(1), 22-28. DOI
- Moran, G.R. (2016) The catalytic function of renalase: A decade of phantoms. Biochim. Biophys. Acta, 1864(1), 177-186. DOI
- Moran, G.R., Hoag, M.R. (2017) The enzyme: Renalase. Arch. Biochem Biophys, 632, 66-76. DOI
- Beaupre, B.A., Hoag, M.R., Roman, J., Forsterling, F.H., Moran, G.R. (2015) Metabolic Function for Human Renalase: Oxidation of Isomeric Forms of beta-NAD(P)H that Are Inhibitory to Primary Metabolism, Biochemistry 54(3), 795-806. DOI
- Wang, Y., Safirstein, R., Velazquez, H., Guo, X.J., Hollander, L., Chang, J., Chen, T.M., Mu, J.J., Desir, G.V. (2017) Extracellular renalase protects cells and organs by outside-in signalling. J. Cell Mol. Med., 21(7), 1260-1265. DOI
- Kolodecik, T.R., Reed, A.M., Date, K., Shugrue, C.A., Patel, V., Chung, S.L., Desir, G.V., Gorelick, F.S. (2017) The serum protein renalase reduces injury in experimental pancreatitis. J. Biol. Chem., 292(51), 21047–21059. DOI
- Wang, L., Velazquez, H., Chang, J., Safirstein, R., Desir, G.V. (2015) Identification of a receptor for extracellular renalase. PLoS One, 10, e0122932. DOI
- Pointer, T.C., Gorelick, F.S., Desir, G.V. (2021) Renalase: A Multi-Functional Signaling Molecule with Roles in Gastrointestinal Disease, Cells. 10, 2006. DOI
- Desir, G.V. (2022) Renalase: discovery, biology, and therapeutic applications. Trans Am Clin Climatol Assoc., 132, 117-125.
- Iwakura, M, Kokubu, T, Ohashi, S. (1993) Immunological application of the dihydrofolate reductase handle carrying a small peptide, leucine enkephalin. J. Biochem. 114(6), 885-889. DOI
- Fedchenko, V.I., Kaloshin, A.A. (2019) A simplified method for obtaining cDNA of low-copy and silent eukaryotic genes using human renalase as an example. Biomedical Chemistry: Research and Methods. 2(2), e00101. DOI
- Fedchenko, V.I., Kaloshin, A.A., Kozlova, N.I., Kopylov, A.T., Medvedev, A.E.(2020) Construction of a chimeric human gene encoding renalase with a modified N-terminus. Biomedical Chemistry: Research and Methods, 3(3), e00137. DOI
- Fedchenko, V.I., Kaloshin, A.A., Kaloshina, S.A., Medvedev, A.E. (2021) Expression and isolation of N-terminal trancated human recombinant renalase in prokariotic cells. Biomedical Chemistry: Research and Methods, 4(3), e00158. DOI
- Laemmli, U.K. (1970) Cleavage of structural proteins during the assemblyof the head of bacteriophage T4. Nature, 227, 680-685. DOI
- Fedchenko, V.I., Veselovsky, A.V., Kopylov, A.T., Kaloshina, S.A., Medvedev, A.E. (2022) Renalase may be cleaved in blood. Are blood chymotrypsin-like enzymes involved?, Medical Hypotheses, 165. 110895, DOI
- Hernández-Preciado M.R., Morán-Moguel, M.C., Dávalos-Rodríguez, I.P., Enríquez-Barajas, C.M., Valdovinos-Maravilla, J.P., Díaz-Pérez, A.L., Silva-Castro, D.E., González-López, L., Gámez-Nava, J.I., Aceves-Aceves, M,A,, Salazar-Páramo, M. (2019) miRNA-24 Gene Sequence, DHFR -829C-T Genotypes, and Methotrexate Response in Mexican Patients with Rheumatoid Arthritis. Genet. Test Mol. Biomarkers. 23(3), 223-227. DOI