Prediction of Peptide Ion Distribution in Positive Electrospray Ionization
Institute of Biomedical Chemistry, 10 Pogodinskaya str., Moscow, 119121 Russia; *e-mail: an.voronina@list.ru
Keywords: peptide; mass-spectrometry; electrospray ionization; property prediction
DOI:10.18097/BMCRM00233
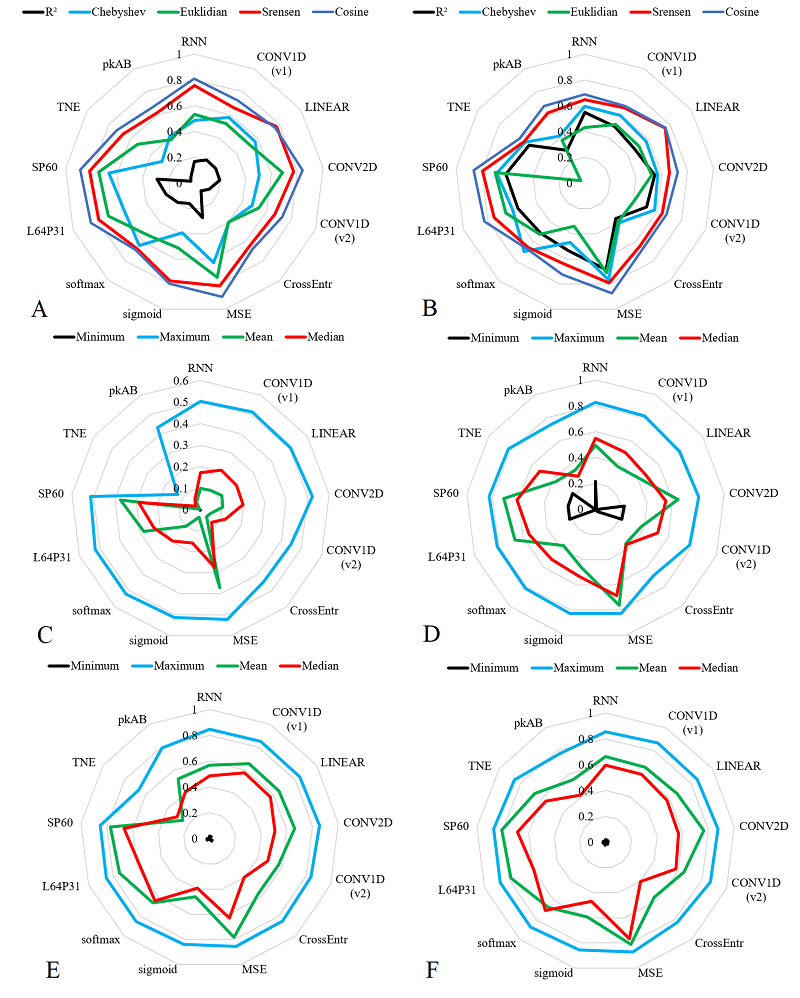
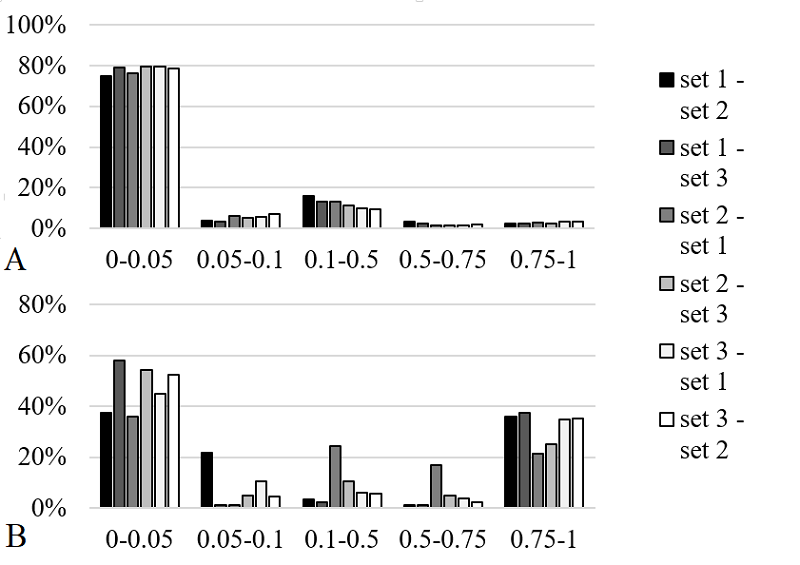
We have investigated the possibility of predicting the distribution of ions of different charge during electrospray ionization of peptides in mass spectrometric experiments using neural networks. Three independent data sets obtained on the same equipment and deposited in ProteomeXchange (PXD032141, PXD051750, PXD019263) were used as training and test samples. A set of fractional values for 1+ to 5+ ions was calculated as predicted values for each of the newly identified peptides. Four different sets of peptide descriptions were used as independent variables, including both the spectrum of amino acid residues and the physicochemical properties of the amino acid residues. Sixty-four variants of neural networks were analyzed, varying the input description, number and type of layers, activation and loss functions. The coefficient of determination and a set of Euclidean, Sorensen, Chebyshev, and Cosine metrics were considered as measures of prediction quality. For the best selected variants, the error did not exceed 10% in 80% of the cases. This accuracy may be sufficient for a preliminary estimation of the probability of detecting a peptide ion of a given charge.
FUNDING
The work was performed within the framework of the Program for Basic Research in the Russian Federation for a long-term period (2021-2030) (No. 122030100170-5).
REFERENCES
- Yates, J.R., Ruse, C.I., Nakorchevsky, A. (2009) Proteomics by mass spectrometry: approaches, advances, and applications. Annu. Rev. Biomed. Eng., 11, 49–79. DOI
- Iavarone, A.T., Jurchen, J.C., Williams, E.R. (2000) Effects of solvent on the maximum charge state and charge state distribution of protein ions produced by electrospray ionization. J. Am. Soc. Mass Spectrom., 11(11), 976–985. DOI
- Skvortsov, V.S., Alekseychuk, N.N., Miroshnichenko, Y.V., Rybina, A.V. (2019) The prediction of the ion fraction of the peptide with selected charge in mass spectrometry with positive electrospray ionization. Biomedical Chemistry: Research and Methods, 2(4), e00100. DOI
- ProteomeXchange. Retrieved July 20, 2024, from: https://proteomecentral.proteomexchange.org
- Ramiro, L., Faura, J., Simats, A., García-Rodríguez, P., Ma, F., Martín, L., Canals, F., Rosell, A., Montaner, J. (2023) Influence of sex, age and diabetes on brain transcriptome and proteome modifications following cerebral ischemia. BMC Neurosci., 24(1), 7. DOI
- Proteomics identification database, project PXD051750. DOI
- Vavilov, N.E., Zgoda, V.G., Tikhonova, O.V., Farafonova, T.E., Shushkova, N.A., Novikova, S.E., Yarygin, K.N., Radko, S.P., Ilgisonis, E.V., Ponomarenko, E.A., Lisitsa, A.V., Archakov, A.I. (2020) Proteomic analysis of Chr 18 proteins using 2D fractionation. J. Proteome Res, 19(12), 4901–4906. DOI
- Voronina, A.I., Miroshnichenko, Yu.V., Skvortsov, V.S. (2024) Bioinformatic identification of proteins with altered PTM levels in a mouse line established to study the mechanisms of the development of fibromuscular dysplasia. Biomeditsinskaya Khimiya, 70(4), 248–255. DOI
- Rybina, A.V. (2024) Identification of proteoforms in experimental ischemic stroke in mice: Comparison of data from 2D electrophoresis and an independent experiment with mass spectrometric identification. Proceedings Book of the XXX Symposium “Bioinformatics and Computer-Aided Drug Discovery”, 116. DOI
- Progenesis LC-MS version 4.0, Nonlinear Dynamics, Newcastle upon Tyne, UK.
- ProteinCNN. Retrieved July 20, 2024, from: https://github.com/rwalroth/ProteinCNN
- Li, Z., Yu, Y. (2016) Protein secondary structure prediction using cascaded convolutional and recurrent neural networks. arXiv preprint 1604.07176. DOI