A Method of Lysate Preparation to Improve the Isolation Efficiency of Protein Partners for Target Proteins Encoded by the Genes of Human Chromosome 18
1Institute of Biomedical Chemistry, , Moscow, 10 Pogodinskaya str., Moscow, 119121 Russian Federation*e-mail: pavel79@inbox.ru
2Institute of Bioorganic Chemistry of the National Academy of Sciences of Belarus, 5, bld. 2 V.F. Kuprevich str., Minsk, 220141 Belarus
Keywords: surface plasmon resonance (SPR); HepG2; lysate preparation; acid treatment and neutralization; protein complexes
DOI:10.18097/BMCRM00090
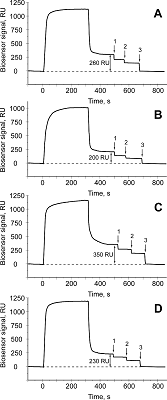
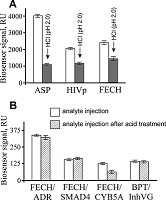
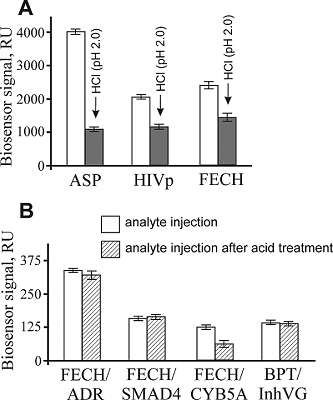
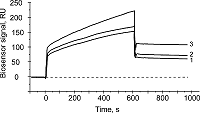
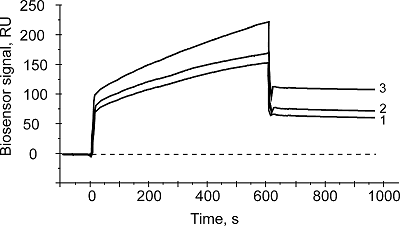
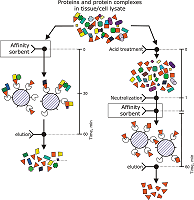
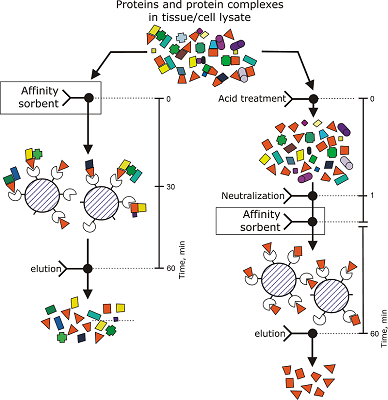
The aim of this work was to test modifications of the standard protocol for the sample preparation of cell/tissue lysate before performing the affinity isolation of lysate protein partners for the target protein (bait protein) which is covalently immobilized on an inert sorbent (e.g. BrCN-, SH-Sepharose 4B) or a carrier (e.g. paramagnetic nanoparticles). The series of our previous works on applying the approach to direct molecular fishing procedure with combination of affinity chromatography and LC-MS/MS analysis using a number of proteins, encoded by the genes of human chromosome 18, have shown that there are at least two problems affecting the specificity and the effectiveness of this procedure. These include: (i) redundancy of the background proteins in the eluates from an affinity sorbent (carrier) due to isolation of multiprotein complexes “labeled” with a direct protein partner which binds with a bait protein immobilized on the sorbent; (ii) low enrichment of the eluates with appropriate protein partners due to the fact that some direct protein partners in the lysate exist in stable “wild type” complexes with the bait protein itself. This means that latter group of protein partners will not be sufficiently isolated from lysate. Therefore, in order to increase the specificity and efficiency of affinity isolation of protein partners for the bait protein, we modified the standard protocol of lysate preparation and the preliminary step on dissociation of lysate protein complexes was added. Several model experiments for the choice of regeneration solution, assessment of their efficiency in the dissociation of lysate protein complexes as well as the stability and binding capacity of proteins were performed under the control of surface plasmon resonance (SPR) biosensor Biacore 3000 using HepG2 cell lysate. It was shown that acid treatment and incubation of the cell lysate for one min on ice (final lysate dilution 20 times) and subsequent neutralization (pH shift from 2.0 to 7.4) resulted in maximal dissociation of the lysate protein complexes without significant negative effects on the protein-protein interactions tested
ACKNOWLEDGEMENTS
The reported study was funded by Russian Foundation for Basic Research (RFBR) аccording to the research project №18-04-00071. Authors used the equipment of “Human Proteome” Core Facility (IBMC, Moscow) supported by Ministry of Education and Science of the Russian Federation (unique ID RFMEFI62117X0017).
REFERENCES
- Ershov, P., Mezentsev, Y., Gnedenko, O., Mukha, D., Yantsevich, A., Britikov, V., Kaluzhskiy, L., Yablokov, E., Molnar, A., Ivanov, A., Lisitsa, A., Gilep, A., Usanov, S., Archakov, A. (2012) Protein interactomics based on direct molecular fishing on paramagnetic particles: experimental simulation and SPR validation. Proteomics, 12(22), 3295-3298. DOI
- Ivanov, A.S., Medvedev, A., Ershov, P., Molnar, A., Mezentsev, Y., Yablokov, E., Kaluzhsky, L., Gnedenko, O., Buneeva, O., Haidukevich, I., Sergeev, G., Lushchyk, A., Yantsevich, A., Medvedeva, M., Kozin, S., Popov, I., Novikova, S., Zgoda, V., Gilep, A., Usanov, S., Lisitsa, A., Archakov, A. (2014) Protein interactomics based on direct molecular fishing on paramagnetic particles: Practical realization and further SPR validation. Proteomics, 14(20), 2261-2274. DOI
- Ivanov, A.S., Ershov, P.V., Molnar, A.A., Mezentsev, Yu.V., Kaluzhskiy, L.A., Yablokov, E.O., Florinskaya, A.V., Gnedenko, O.V., Medvedev, A.E., Kozin, S.A., Mitkevich, V.A., Makarov, A.A., Gilep, A.A., Luschik, A.Ya., Gaidukevich, I.V., Usanov S.A. (2016) Direct molecular fishing in molecular partners investigation in protein–protein and protein–peptide interactions. RJBC, 42(1), 14-21. DOI
- Svirid, A.V., Ershov, P.V., Yablokov, E.O., Kaluzhskiy, L.A., Mezentsev, Y.V., Florinskaya, A.V., Sushko, T.A., Strushkevich, N.V., Gilep, A.A., Usanov, S.A., Medvedev, A.E., Ivanov, A.S. (2017) Direct molecular fishing of new protein partners for human thromboxane synthase. Acta Naturae. 9(4), 92-100.
- Florinskaya, A., Ershov, P., Mezentsev, Y., Kaluzhskiy, L., Yablokov, E., Medvedev, A., Ivanov, A. (2018) SPR Biosensors in Direct Molecular Fishing: Implications for Protein Interactomics. Sensors (Basel), 18(5), 1616. DOI
- Gilep, A.A., Guryev. O.L., Usanov, S.A., Estabrook, R.W. (2001) Apo-cytochrome b5 as an indicator of changes in heme accessability: preliminary studies with cytochrome P450 3A4. J. Inorg. Biochem., 87(4), 237-244. DOI
- Usanov, S.A., Graham, S.E., Lepesheva, G.I., Azeva, T.N., Strushkevich, N.V., Gilep, A.A., Estabrook, R.W., Peterson, J.A. (2002) Probing the interaction of bovine cytochrome P450scc (CYP11A1) with adrenodoxin: evaluating site-directed mutations by molecular modeling. Biochemistry, 41(26), 8310–8320. DOI
- Sergeev, G.V., Gilep, A.A. & Usanov S.A. (2014) The role of cytochrome b5 structural domains in interaction with cytochromes P450. Biochemistry (Moscow), 79(5), 406-416. DOI
- Naryzhny, S.N., Maynskova, M.A., Zgoda, V.G., Ronzhina, N.L., Kleyst, O.A., Vakhrushev, I.V., Archakov, A.I. (2016) Virtual-Experimental 2DE Approach in Chromosome-Centric Human Proteome Project. J. Proteome Res., 15(2), 525–530. DOI
- Mezentsev, Yu.V., Molnar, A.A., Gnedenko, O.V., Krasotkina, Yu.V., Sokolov, N.N., Ivanov, A.S. (2006) Oligomerization of L-asparaginase from Erwinia carotovora. Biochem. Moscow Suppl. Ser. B, 1(1), 58-67. DOI
- Mezentsev, Yu.V., Molnar, A.A., Sokolov, N.N., Lisitsina, V.B., Shatskaya, M.A., Ivanov, A.S., Archakov, A.I. (2011) Specificity of Molecular Recognition in Oligomerization of Bacterial L-Asparaginases. Biochem. Moscow Suppl. Ser. B, 5(2), 124-134. DOI
- Ershov, P.V., Gnedenko, O.V., Mol'nar, A.A., Lisitsa, A.V., Ivanov, A.S., Archakov, A.I. (2009) Biosensor analysis of interaction of potential dimerization inhibitors with HIV-1 protease. Biochem. Moscow Suppl. Ser. B, 3(3), 272-288. DOI
- Ershov, P., Mezentsev, Y., Gilep, A., Usanov, S., Buneeva, O., Medvedev, A., Ivanov, A. (2017) Isatin-induced increase in the affinity of human ferrochelatase and adrenodoxin reductase interaction. Protein Sci., 26(12), 2458-2462. DOI
- Fleischmann, G., Fisette, O., Thomas, C., Wieneke, R., Tumulka, F., Schneeweiss, C., Springer, S., Schäfer, L.V., Tampé, R. (2015) Mechanistic Basis for Epitope Proofreading in the Peptide-Loading Complex. J. Immunol., 195(9), 4503-4513. DOI