Evaluation of the Collateral Activity of CRISPR/Cas13a-Ribonuclease on the Agilent 2100 Bioanalyzer
Institute of Biomedical Chemistry, 10 Pogodinskaya str., Moscow, 119121 Russia; *e-mail: radkos@yandex.ru
Keywords: CRISPR/Cas13a-ribonuclease; collateral activity; detection; Agilent 2100 bioanalyzer
DOI:10.18097/BMCRM00169
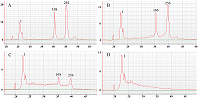
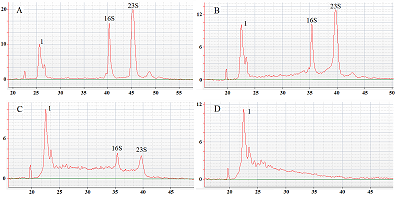
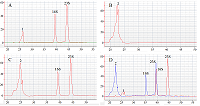
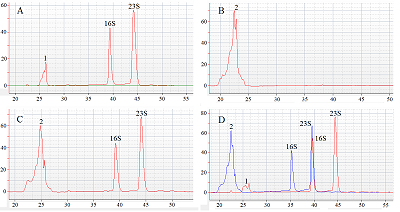
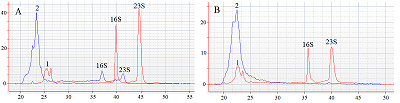
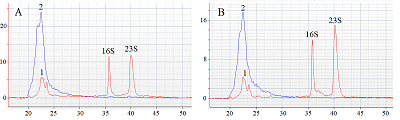
The paper describes an original technique for detecting the collateral activity of CRISPR/Cas 13a ribonuclease based on the assessment of ribosomal RNA degradation. The Agilent 2100 bioanalyzer is used as an analyzing device. This approach is an alternative to existing detection methods and has a number of advantages over them in the case when a quantitative assessment of activity is not required. On the example of the test sample, the optimal concentrations and ratios of the components of the reaction mixture, which are necessary to obtain the most indicative result, were determined. The proposed technique can be used for qualitative assessment of the activity of recombinant ribonuclease Cas13a preparations obtained under different conditions of heterologous protein expression and purification, as well as for testing guide RNAs.
FUNDING
The study was supported by the Program of Fundamental Research in the Russian Federation for a Long Term of 2021–2030.
REFERENCES
- Gootenberg, J.S., Abudayyeh, O.O., Lee, J.W., Essletzbichler, P., Dy, A.J., Joung, J., Verdine, V., Donghia, N., Daringer, N.M., Freije, C.A., Myhrvold, C., Bhattacharyya, R.P., Livny, J., Regev, A., Koonin, E.V., Hung, D.T., Sabeti, P.C., Collins, J.J., Zhang, F. (2017) Nucleic acid detection with CRISPR-Cas13a/C2c2. Science, 356(6336), 438-442. DOI
- Gootenberg, J.S., Abudayyeh, O.O., Kellner, M.J., Joung, J., Collins, J.J., Zhang, F. (2018) Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science, 360(6387), 439-444. DOI
- Kellner, M.J., Koob, J.G., Gootenberg, J.S., Abudayyeh, O.O., Zhang, F. (2019) SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat. Protoc., 14(10), 2986-3012. DOI
- Myhrvold, C., Freije, C.A., Gootenberg, J.S., Abudayyeh, O.O., Metsky, H.C., Durbin, A.F., Kellner, M.J., Tan, A.L., Paul, L.M., Parham, L.A., Garcia, K.F., Barnes, K.G., Chak, B., Mondini, A., Nogueira, M.L., Isern, S., Michael, S.F., Lorenzana, I., Yozwiak, N.L., MacInnis, B.L., Bosch, I., Gehrke, L., Zhang, F., Sabeti, P.C. (2018) Field-deployable viral diagnostics using CRISPR-Cas13. Science, 360(6387), 444-448. DOI
- Li, Y., Li, S., Wang, J., Liu, G. (2019) CRISPR/Cas Systems towards Next-Generation Biosensing. Trends Biotechnol., 37(7), 730-743. DOI
- Kurbatov, L.K., Radko, P.S., Kravchenko, S.V., Kiseleva, O.I., Durmanov, N.D., Lisitsa, A.V. (2020) Single Stage Purification of CRISPR/Cas13a Nuclease by Metal-Chelating Chromatography Following Heterologous Expression with Preservation of Collateral Ribonuclease Activity. Appl. Biochem. Microbiol., 56(6), 671-677. DOI
- Thormann, W. (2018) Theoretical principles of capillary electromigration methods., in Capillary Electromigration Separation Methods, Poole, C. F., Editor, Elsevier. p. 21-44.